








生物技术通报 ›› 2021, Vol. 37 ›› Issue (12): 60-70.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0141
韩占红1( ), 宗元元1, 张学梅1, 王斌1, PRUSKY Dov2, 毕阳1(
), 宗元元1, 张学梅1, 王斌1, PRUSKY Dov2, 毕阳1( )
)
收稿日期:2021-02-04
出版日期:2021-12-26
发布日期:2022-01-19
作者简介:韩占红,硕士研究生,研究方向:采后生物学与技术;E-mail: 基金资助:
HAN Zhan-hong1( ), ZONG Yuan-yuan1, ZHANG Xue-mei1, WANG Bin1, PRUSKY Dov2, BI Yang1(
), ZONG Yuan-yuan1, ZHANG Xue-mei1, WANG Bin1, PRUSKY Dov2, BI Yang1( )
)
Received:2021-02-04
Published:2021-12-26
Online:2022-01-19
摘要:
麦角甾醇是真菌细胞质膜的特有组分,在真菌生长发育中具有重要作用。erg4是参与麦角甾醇生物合成最后一步反应的基因,但扩展青霉中该基因的功能未知。本文通过RT-PCR方法克隆了扩展青霉3个erg4(erg4A、erg4B 和 erg4C)基因的 CDS全长,对基因结构、编码蛋白的跨膜螺旋和亲疏水性进行了生物信息学分析,通过融合绿色荧光蛋白定位的方法进行了亚细胞定位,测定了3个基因在不同生长发育阶段、不同培养基状态以及黑暗和蓝光条件下的表达差异。扩展青霉erg4A、eerg4B 和 erg4C的 CDS全长分别为1 476 bp、1 491 bp和 1 596 bp,分别编码491、496和 531个氨基酸;编码蛋白均属于跨膜蛋白,且表现出疏水性。绿色荧光蛋白与内质网红色荧光探针染色共定位结果显示,Erg4A、Erg4B和Erg4C均定位于内质网。erg4A、erg4B 和erg4C在孢子阶段、孢子萌发阶段及成熟菌丝阶段的表达水平存在显著差异,其中,erg4A在3个阶段的表达量无明显变化,而erg4B和 erg4C的表达量均显著上调,以erg4B的上调幅度最为明显。erg4A在 CY液体和固体培养条件下的表达量无显著变化,erg4B和erg4C在CY液体培养条件下的表达量显著高于固体培养,以erg4B的上调幅度最为明显。erg4A和 erg4B在蓝光条件下的表达量显著高于黑暗条件,以erg4B的上调幅度最为明显。erg4C对蓝光条件不敏感。扩展青霉Erg4A、Erg4B和 Erg4C均定位于内质网,erg4A、erg4B和 erg4C在不同生长发育阶段、固体和液体以及黑暗与蓝光培养条件下的表达存在较大差异,其中,以erg4B的响应最为活跃。
韩占红, 宗元元, 张学梅, 王斌, PRUSKY Dov, 毕阳. 扩展青霉erg4的生物信息学、亚细胞定位及表达分析[J]. 生物技术通报, 2021, 37(12): 60-70.
HAN Zhan-hong, ZONG Yuan-yuan, ZHANG Xue-mei, WANG Bin, PRUSKY Dov, BI Yang. Bioinformatics,Subcellular Localization and Expression Analysis of erg4 in Penicillium expansum[J]. Biotechnology Bulletin, 2021, 37(12): 60-70.
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAAATTGCAGTAG | |
| erg4B | F:ATGCCCTCCAAAAAGGACTCTC |
| R:GTAAATTCCAGGAATGATGC | |
| erg4C | F:ATGGATCGTCCCGGCTTCATT |
| R:TTAGAAGACATAAGGAATGA |
表1 erg4A、erg4B和erg4C基因CDS的克隆引物序列
Table 1 Primers sequence for the clone of CDS of gene erg4A,erg4B and erg4C
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAAATTGCAGTAG | |
| erg4B | F:ATGCCCTCCAAAAAGGACTCTC |
| R:GTAAATTCCAGGAATGATGC | |
| erg4C | F:ATGGATCGTCCCGGCTTCATT |
| R:TTAGAAGACATAAGGAATGA |
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | CF:TCAACTCCATCACATCACAAGAGCTC ATGGAGG- ACC AAGTCAAATC |
| CR:AGCTCCTCGCCCTTGCTCACTCTAGAAAAAAGAG- AA ATTGCAGTAG | |
| erg4B | CF:TCAACTCCATCACATCACAA GAGCTC ATGCCCTC- CA AAAAGGACTC |
| CR:AGCTCCTCGCCCTTGCTCAC TCTAGA GTAAATTC- CA GGAATGATGC | |
| erg4C | NF:GCATGGACGAGCTGTACAAG GAGCTC ATGGATC- GTCCCGGCTTCAT |
| NR:ATGGAGCTATTAAATCACTA TCTAGA TTAGAAG- ACA TACTATAGGA |
表2 用于构建GFP融合表达载体的引物序列
Table 2 Primer sequences for GFP fusion vector construction
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | CF:TCAACTCCATCACATCACAAGAGCTC ATGGAGG- ACC AAGTCAAATC |
| CR:AGCTCCTCGCCCTTGCTCACTCTAGAAAAAAGAG- AA ATTGCAGTAG | |
| erg4B | CF:TCAACTCCATCACATCACAA GAGCTC ATGCCCTC- CA AAAAGGACTC |
| CR:AGCTCCTCGCCCTTGCTCAC TCTAGA GTAAATTC- CA GGAATGATGC | |
| erg4C | NF:GCATGGACGAGCTGTACAAG GAGCTC ATGGATC- GTCCCGGCTTCAT |
| NR:ATGGAGCTATTAAATCACTA TCTAGA TTAGAAG- ACA TACTATAGGA |
| Gene | Primer sequence(5' - 3') |
|---|---|
| C-CX | CX1:GGAGA CGTAT TTAGGTGCTA |
| CX2:TGAACTTCA GGGTCAGCTT | |
| N-CX | NX1:CGACAACCAC TACCTGAGCA |
| NX2:TGAAGGGCGT ACTAGGGTTG | |
| erg4A-GFP | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAA ATTGCAGTAG | |
| erg4B-GFP | F:ATGCCCTCCAAAAAGGACTC |
| R:GTAAATTCCA GGAATGATGC | |
| erg4C-GFP | F:ATGGATCGTC CCGGCTTCAT |
| R:TTAGAAGACA TAAGGAATGA |
表3 转化子验证所用引物序列
Table 3 Primer sequences for transformant validation
| Gene | Primer sequence(5' - 3') |
|---|---|
| C-CX | CX1:GGAGA CGTAT TTAGGTGCTA |
| CX2:TGAACTTCA GGGTCAGCTT | |
| N-CX | NX1:CGACAACCAC TACCTGAGCA |
| NX2:TGAAGGGCGT ACTAGGGTTG | |
| erg4A-GFP | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAA ATTGCAGTAG | |
| erg4B-GFP | F:ATGCCCTCCAAAAAGGACTC |
| R:GTAAATTCCA GGAATGATGC | |
| erg4C-GFP | F:ATGGATCGTC CCGGCTTCAT |
| R:TTAGAAGACA TAAGGAATGA |
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:TCCACGAATGTTCGGAATCC |
| R:AGCTCAAACCCAAGAGCATGA | |
| erg4B | F:CGGCGCTTTATGATACCAATG |
| R:GGGAAGGAAGACCTGCAAAAA | |
| erg4C | F:GCATTCCCATCAATCAAAGCA |
| R:CCGGGAAGAAGCACGTAACA | |
| β-tubulin | F:CTCCAGCTCGAGCGTATGAAC |
| R:GGCTCCAAATCGACGAGAAC |
表4 实时荧光定量PCR引物序列
Table 4 Primers for real-time fluorescent quantitative PCR
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:TCCACGAATGTTCGGAATCC |
| R:AGCTCAAACCCAAGAGCATGA | |
| erg4B | F:CGGCGCTTTATGATACCAATG |
| R:GGGAAGGAAGACCTGCAAAAA | |
| erg4C | F:GCATTCCCATCAATCAAAGCA |
| R:CCGGGAAGAAGCACGTAACA | |
| β-tubulin | F:CTCCAGCTCGAGCGTATGAAC |
| R:GGCTCCAAATCGACGAGAAC |
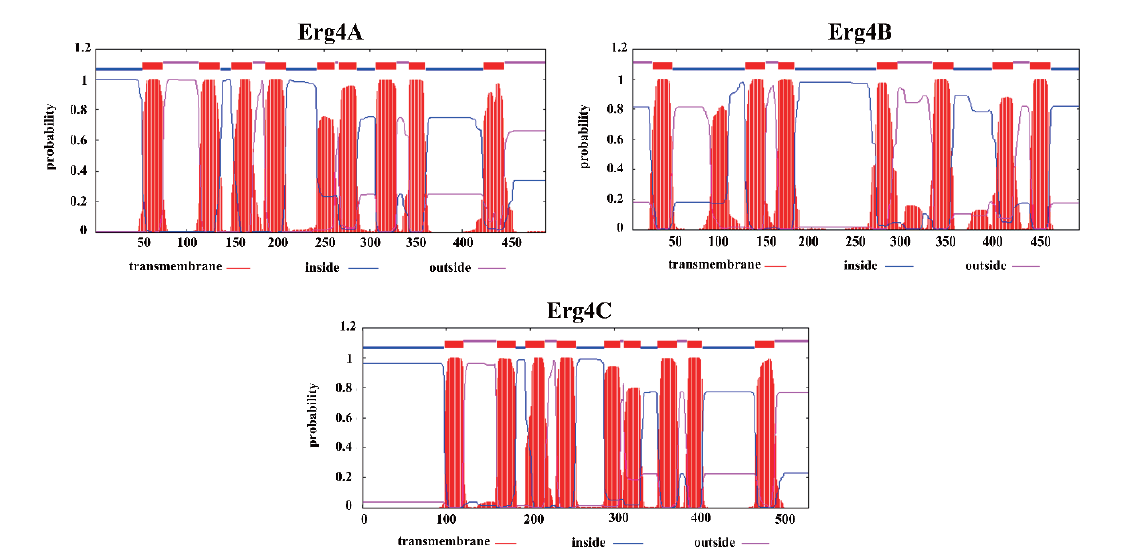
图3 Erg4A、Erg4B和Erg4C的跨膜螺旋 图中的红色矩形为跨膜螺旋结构,蓝色线段是位于膜内的结构,而红色线段是位于膜外的结构
Fig. 3 Transmembrane helices of Erg4A,Erg4B and Erg4C Red rectangle indicates the transmembrane,blue line segment indicates the structure inside the membrane,and the red line segment indicates the structure outside the membrane

图4 Erg4A、Erg4B和Erg4C蛋白亲疏水性 图中的正值区域代表疏水氨基酸,负值区域代表亲水氨基酸
Fig. 4 Hydrophilicity/Hydrophobicity of Erg4A,Erg4B and Erg4C protein Positive areas represent the hydrophobic,the negative areas represent the hydrophilic
图6 野生型与荧光标记菌株erg4A-GFP、erg4B-GFP及erg4C-GFP的菌落形态比较
Fig. 6 Comparison of colony morphologies of erg4A-GFP,erg4B-GFP and erg4C-GFP in wild type and fluor-esence labelled strains
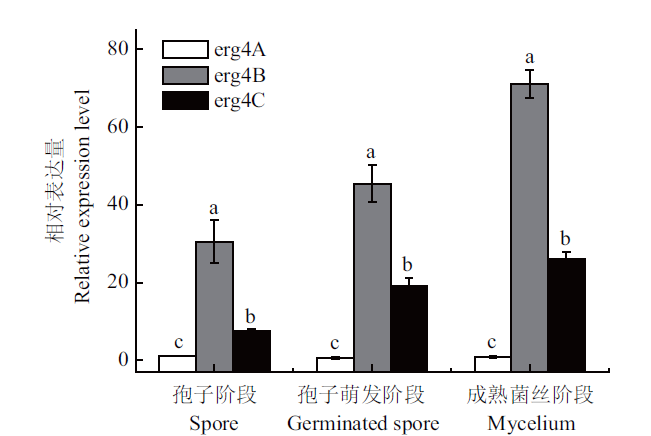
图10 扩展青霉中erg4A、erg4B和erg4C在孢子阶段、孢子萌发阶段以及成熟菌丝阶段的相对表达量 竖线表示标准误(±SE). 不同字母代表显著性差异(P<0.05),下同
Fig. 10 Relative expressions of erg4A,erg4B and erg4C in the spore,germinated spore and mycelium stages in P. expansum Vertical line indicates standard error(±SE). Different letters refer to significant differences(P<0.05). The same below
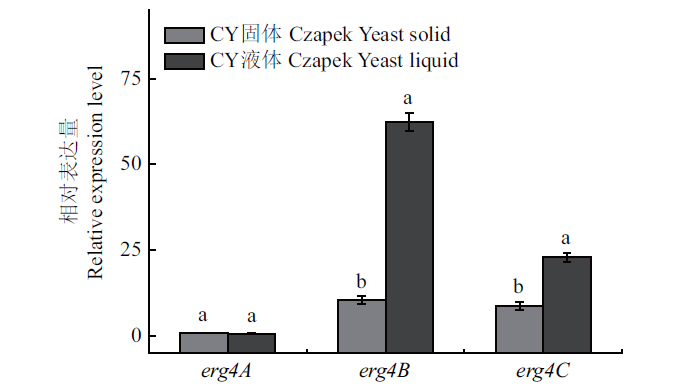
图11 扩展青霉中erg4A、erg4B和erg4C在CY固体和液体培养条件下的相对表达量
Fig. 11 Relative expressions of erg4A,erg4B and erg4C in P. expansum in Czapek yeast extract solid medium and Czapek yeast extract liquid medium
| [1] |
Sun C, Fu D, Lu HP, et al. Autoclaved yeast enhances the resistance against Penicillium expansum in postharvest pear fruit and its possible mechanisms of action[J]. Biological Control, 2018, 119:51-58.
doi: 10.1016/j.biocontrol.2018.01.010 URL |
| [2] |
Nierop Groot M, Abee T, van Bokhorst-van de Veen H. Inactivation of conidia from three Penicillium spp. isolated from fruit juices by conventional and alternative mild preservation technologies and disinfection treatments[J]. Food Microbiol, 2019, 81:108-114.
doi: S0740-0020(17)31140-1 pmid: 30910081 |
| [3] |
Hu Z, He B, Ma L, et al. Recent advances in ergosterol biosynjournal and regulation mechanisms in Saccharomyces cerevisiae[J]. Indian J Microbiol, 2017, 57(3):270-277.
doi: 10.1007/s12088-017-0657-1 URL |
| [4] | Sun L, Liao K. The effect of honokiol on ergosterol biosynjournal and vacuole function in Candida albicans[J]. Journal of Microbiol Biotechnology and biotechnology, 2020, 30(12):1835-1842. |
| [5] |
Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast[J]. J Cell Biol, 2008, 180(4):813-826.
doi: 10.1083/jcb.200705076 URL |
| [6] | Song J, Zhai P, Zhang Y, et al. The Aspergillus fumigatus damage resistance protein family coordinately regulates ergosterol biosynjournal and azole susceptibility[J]. mBio, 2016, 7(1):e01919-e01915. |
| [7] |
Alcazar-Fuoli L, Mellado E. Ergosterol biosynjournal in Aspergillus fumigatus:its relevance as an antifungal target and role in antifungal drug resistance[J]. Front Microbiol, 2012, 3:439.
doi: 10.3389/fmicb.2012.00439 pmid: 23335918 |
| [8] |
Liu JF, Xia JJ, Nie KL, et al. Outline of the biosynjournal and regulation of ergosterol in yeast[J]. World J Microbiol Biotechnol, 2019, 35(7):98.
doi: 10.1007/s11274-019-2673-2 URL |
| [9] |
Yun Y, Yin D, Dawood DH, et al. Functional characterization of FgERG3 and FgERG5 associated with ergosterol biosynjournal, vegetative differentiation and virulence of Fusarium graminearum[J]. Fungal Genet Biol, 2014, 68:60-70.
doi: 10.1016/j.fgb.2014.04.010 URL |
| [10] |
Jordá T, Puig S. Regulation of ergosterol biosynjournal in Saccharomyces cerevisiae[J]. Genes, 2020, 11(7):795.
doi: 10.3390/genes11070795 URL |
| [11] |
Zweytick D, Hrastnik C, Kohlwein SD, et al. Biochemical characterization and subcellular localization of the sterol C-24(28)reductase, Erg4p, from the yeastSaccharomyces cerevisiae[J]. FEBS Lett, 2000, 470(1):83-87.
pmid: 10722850 |
| [12] |
Feng W, Yang J, Xi Z, et al. Mutations and/or overexpressions of ERG4 and ERG11 genes in clinical azoles-resistant isolates of Candida albicans[J]. Microb Drug Resist, 2017, 23(5):563-570.
doi: 10.1089/mdr.2016.0095 URL |
| [13] |
Tiedje C, Holland DG, Just U, et al. Proteins involved in sterol synjournal interact with Ste20 and regulate cell polarity[J]. J Cell Sci, 2007, 120(pt 20):3613-3624.
doi: 10.1242/jcs.009860 URL |
| [14] |
Aguilar PS, Heiman MG, Walther TC, et al. Structure of sterol aliphatic chains affects yeast cell shape and cell fusion during mating[J]. PNAS, 2010, 107(9):4170-4175.
doi: 10.1073/pnas.0914094107 pmid: 20150508 |
| [15] | Bhattacharya S, Esquivel BD, White TC. Overexpression or deletion of ergosterol biosynjournal genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae[J]. mBio, 2018, 9(4):e01291-e01218. |
| [16] |
Venegas M, Barahona S, González AM, et al. Phenotypic analysis of mutants of ergosterol biosynjournal genes(ERG3 and ERG4)in the red yeast Xanthophyllomyces dendrorhous[J]. Front Microbiol, 2020, 11:1312.
doi: 10.3389/fmicb.2020.01312 URL |
| [17] |
Liu X, Jiang JH, Jiang JH, et al. Sterol C-24(28)reductase, encoded by FgERG4 gene, is important for ergosterol production, hyphal growth, sporulation and virulence in Fusarium graminearum[J]. Molecular plant pathology, 2013, 14(1):71-83.
doi: 10.1111/mpp.2013.14.issue-1 URL |
| [18] | Long NB, Xu XL, Zeng QQ, et al. Erg4A and Erg4B are required for conidiation and azole resistance via regulation of ergosterol biosynjournal in Aspergillus fumigatus[J]. Appl Environ Microbiol, 2017, 83(4):e02924-e02916. |
| [19] |
Li B, Zong Y, Du Z, et al. Genomic characterization reveals insights into patulin biosynjournal and pathogenicity in Penicillium species[J]. Mol Plant Microbe Interact, 2015, 28(6):635-647.
doi: 10.1094/MPMI-12-14-0398-FI URL |
| [20] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
pmid: 11846609 |
| [21] |
Tahir II, Department of Plant Breeding Swedish University of Agricultural Sciences Sweden, Nybom H, et al. Susceptibility to blue mold caused by Penicillium expansum in apple cultivars adapted to a cool climate[J]. Europ J Hort Sci, 2015, 80(3):117-127.
doi: 10.17660/eJHS.2015/80.3.4 URL |
| [22] | 刘馨. 小麦赤霉病菌麦角甾醇生物合成途径中关键基因的功能研究[D]. 杭州:浙江大学, 2012. |
| Liu X. Functional analysis of genes involved in the ergosterol biosynthesis pathway in Fusarium graminearum[D]. Hangzhou:Zhejiang University, 2012. | |
| [23] |
Manchalu S, Mittal N, Spang A, et al. Local translation of yeast ERG4 mRNA at the endoplasmic Reticulum requires the brefeldin A resistance protein Bfr1[J]. RNA, 2019, 25(12):1661-1672.
doi: 10.1261/rna.072017.119 URL |
| [24] | 徐孝苓. 烟曲霉麦角固醇合成相关基因erg4的功能性研究[D]. 南京:南京师范大学, 2017. |
| Xu XL. Functional analysis of ergosterol biosynthesis related gene erg4 in Aspergillus fumigatus[D]. Nanjing:Nanjing Normal University, 2017. | |
| [25] | 于忠洋, 姜洋, 翟明霞, 等. 桑黄麦角甾醇生物合成关键酶PlERG24基因克隆与表达分析[J]. 中草药, 2020, 51(22):5825-5832. |
| Yu ZY, Jiang Y, Zhai MX, et al. Cloning and expression analysis of PlERG24 gene in ergosterol biosynjournal of Phellinus linteus[J]. Chin Tradit Herb Drugs, 2020, 51(22):5825-5832. | |
| [26] |
Rosenfeld E, Beauvoit B. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae[J]. Yeast, 2003, 20(13):1115-1144.
pmid: 14558145 |
| [27] |
Tang Y, Zhu PK, Lu Z, et al. The photoreceptor components FaWC1 and FaWC2 of fusarium asiaticum cooperatively regulate light responses but play independent roles in virulence expression[J]. Microorganisms, 2020, 8(3):365.
doi: 10.3390/microorganisms8030365 URL |
| [28] |
Casas-Flores S, Rios-Momberg M, Bibbins M, et al. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride[J]. Microbiol Read Engl, 2004, 150(Pt 11):3561-3569.
doi: 10.1099/mic.0.27346-0 URL |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [3] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [4] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [5] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [6] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [7] | 于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193. |
| [8] | 陈佳敏, 刘永杰, 马锦绣, 李丹, 公杰, 赵昌平, 耿洪伟, 高世庆. 小麦组蛋白甲基化酶在杂交种中干旱胁迫表达模式分析[J]. 生物技术通报, 2022, 38(7): 51-61. |
| [9] | 杨佳宝, 周至铭, 张展, 冯丽, 孙黎. 向日葵HaLACS1的克隆、表达及酵母功能互补鉴定[J]. 生物技术通报, 2022, 38(6): 147-156. |
| [10] | 王楠, 张瑞, 潘阳阳, 何翃宏, 王靖雷, 崔燕, 余四九. 牦牛TGF-β1基因克隆及在雌性生殖系统主要器官中的表达定位[J]. 生物技术通报, 2022, 38(6): 279-290. |
| [11] | 李宇航, 王兴平, 杨箭, 罗仍卓么, 任倩倩, 魏大为, 马云. miR-665在奶牛乳腺上皮细胞炎症中的表达及功能分析[J]. 生物技术通报, 2022, 38(5): 159-168. |
| [12] | 李洋, 张晓天, 朴静子, 周如军, 李自博, 关海雯. 花生疮痂病菌蓝光受体EaWC 1基因克隆及生物信息学分析[J]. 生物技术通报, 2022, 38(5): 93-99. |
| [13] | 镐青青, 姚圣, 刘佳禾, 陈佩珍, 张梦洋, 季孔庶. 马尾松NAC转录因子基因PmNAC8的克隆及表达分析[J]. 生物技术通报, 2022, 38(4): 202-216. |
| [14] | 赵婷婷, 王俊刚, 王文治, 冯翠莲, 冯小艳, 张树珍. 甘蔗单糖转运蛋白基因ShSTP7序列分析及组织表达特征测定[J]. 生物技术通报, 2022, 38(4): 72-78. |
| [15] | 张琳, 魏祯祯, 宋程威, 郭丽丽, 郭琪, 侯小改, 王华芳. ‘凤丹’牡丹PoFD基因克隆及表达分析[J]. 生物技术通报, 2022, 38(11): 104-111. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||