








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 42-56.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1158
• 食用菌生物技术专题(专题主编: 黄晨阳) • 上一篇 下一篇
收稿日期:2021-09-09
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:陈体强,男,研究员,研究方向:药用真菌;E-mail: 基金资助:
CHEN Ti-qiang1( ), XU Xiao-lan2, SHI Lin-chun3, ZHONG Li-Yi4
), XU Xiao-lan2, SHI Lin-chun3, ZHONG Li-Yi4
Received:2021-09-09
Published:2021-11-26
Online:2021-12-03
摘要:
采用 PacBio SMART技术并基于三代和二代数据(3+2策略)完成紫芝栽培品种‘武芝2号’(原名:S2)(通称‘紫芝S2’)基因组De novo测序、组装,完整地获得紫芝栽培品种‘武芝2号’(原名:S2)全基因组,序列总长度56.849 Mb,包括核基因组和线粒体基因组共72条Scaffolds。其中,核基因组16 681个基因,完成其中13 161个基因的注释,约占总数的78.89%;预测分析到4组串联重复的rRNA簇、405个非编码RNA 和3 305个启动子,并找到rpb2、beta-tubulin和18S,28S rRNA(SSU,LSU)等基因序列以及漆酶同工酶基因,鲨烯合酶、羊毛甾醇合酶基因与麦角甾醇合成代谢相关基因及其分布位置,为今后紫芝栽培品种(系)分子鉴定与菌株鉴别、功能基因的挖掘与利用提供可靠信息,同时也为其后续开展中国栽培灵芝种类的比较基因组学分析积累科学数据。
陈体强, 徐晓兰, 石林春, 钟礼义. 紫芝栽培品种‘武芝2号’(‘紫芝S2’)全基因组测序及分析[J]. 生物技术通报, 2021, 37(11): 42-56.
CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2)[J]. Biotechnology Bulletin, 2021, 37(11): 42-56.
| 数据Data | Read总条数 Total number of reads | 碱基总数目 Total base/bp | Reads长度 Reads length/bp | GC% | Q20* | Q30** |
|---|---|---|---|---|---|---|
| Raw data | 61 457 184 | 9 218 577 600 | 150∶150 | 54.37 | 97.49 | 93.09 |
| Clean data | 55 973 534 | 8 396 030 100 | 150∶150 | 54.36 | 97.33 | 92.81 |
表1 二代测序(Illumina Nova Seq6000)数据统计及过滤结果
Table 1 Statistics of second generation sequencing data(Illumina Nova Seq6000)and filtering results
| 数据Data | Read总条数 Total number of reads | 碱基总数目 Total base/bp | Reads长度 Reads length/bp | GC% | Q20* | Q30** |
|---|---|---|---|---|---|---|
| Raw data | 61 457 184 | 9 218 577 600 | 150∶150 | 54.37 | 97.49 | 93.09 |
| Clean data | 55 973 534 | 8 396 030 100 | 150∶150 | 54.36 | 97.33 | 92.81 |
| 项目 Item | 数据 Data | |
|---|---|---|
| 下机的碱基数目 | Polymerase read total bases(bp) | 11 049 130 528 |
| 下机的reads数目 | Number of polymerase reads | 531 175 |
| 下机的read平均长度 | Post-filter mean read length | 20 801 |
| 下机的read的N50长度 | Polymerase read N50 | 35 250 |
| 去掉接头后的碱基数目 | Subreads total bases(bp) | 11 029 483 296 |
| 去掉接头后的 Subread数目 | Number of subreads | 970 426 |
| 去掉接头后的 Subread 平均长度 | Mean subread length(bp) | 11 365.61 |
| 去掉接头后的 Subread N50长度 | Subreads N50 length | 15 985 |
| 过滤无效数据后Subreads总长度 | Subreads total bases(bp) | 10 997 790 898 |
| 过滤无效数据后Subreads数目 | Number of subreads | 908 820 |
| 过滤无效数据后Subreads平均长度* | Subreads mean length(bp)* | 12 101 |
| 过滤无效数据后Subreads N50长度 | Subreads N50 length(bp) | 16 018* |
表2 三代测序(PacBio Sequel)下机数据统计及过滤结果
Table 2 Statistics of third generation sequencing data and filtering results
| 项目 Item | 数据 Data | |
|---|---|---|
| 下机的碱基数目 | Polymerase read total bases(bp) | 11 049 130 528 |
| 下机的reads数目 | Number of polymerase reads | 531 175 |
| 下机的read平均长度 | Post-filter mean read length | 20 801 |
| 下机的read的N50长度 | Polymerase read N50 | 35 250 |
| 去掉接头后的碱基数目 | Subreads total bases(bp) | 11 029 483 296 |
| 去掉接头后的 Subread数目 | Number of subreads | 970 426 |
| 去掉接头后的 Subread 平均长度 | Mean subread length(bp) | 11 365.61 |
| 去掉接头后的 Subread N50长度 | Subreads N50 length | 15 985 |
| 过滤无效数据后Subreads总长度 | Subreads total bases(bp) | 10 997 790 898 |
| 过滤无效数据后Subreads数目 | Number of subreads | 908 820 |
| 过滤无效数据后Subreads平均长度* | Subreads mean length(bp)* | 12 101 |
| 过滤无效数据后Subreads N50长度 | Subreads N50 length(bp) | 16 018* |
| Property | Min | Max | Min | Max | Min | Max |
|---|---|---|---|---|---|---|
| Heterozygosity/% | 1.965 62 | 2.008 05 | 1.955 13 | 1.973 2 | 1.958 25 | 1.984 25 |
| Genome haploid length/ bp | 46 347 904 | 46 440 807 | 50 222 379 | 50 301 292 | 48 878 189 | 48 958 199 |
| Genome repeat length/ bp | 4 637 105 | 4 646 400 | 8 024 318 | 8 036 926 | 6 841 040 | 6 852 239 |
| Sequences repeat rate/% | 10.005 | 10.005 | 15.977 | 15.977 | 13.996 | 13.996 |
| Genome unique length/ bp | 41 710 799 | 41 794 407 | 42 198 061 | 42 264 366 | 42 037 149 | 42 105 960 |
| Model fit/% | 95.5952 | NA | 96.9219 | 98.2063 | 96.2289 | 96.9645 |
| Read error rate/% | 0.253032 | 0.253032 | 0.229964 | 0.229964 | 0.238092 | 0.238092 |
表3 基因组大小与杂合度评估(k = 17)
Table 3 Assessment of genome size and heterozygosity(k = 17)
| Property | Min | Max | Min | Max | Min | Max |
|---|---|---|---|---|---|---|
| Heterozygosity/% | 1.965 62 | 2.008 05 | 1.955 13 | 1.973 2 | 1.958 25 | 1.984 25 |
| Genome haploid length/ bp | 46 347 904 | 46 440 807 | 50 222 379 | 50 301 292 | 48 878 189 | 48 958 199 |
| Genome repeat length/ bp | 4 637 105 | 4 646 400 | 8 024 318 | 8 036 926 | 6 841 040 | 6 852 239 |
| Sequences repeat rate/% | 10.005 | 10.005 | 15.977 | 15.977 | 13.996 | 13.996 |
| Genome unique length/ bp | 41 710 799 | 41 794 407 | 42 198 061 | 42 264 366 | 42 037 149 | 42 105 960 |
| Model fit/% | 95.5952 | NA | 96.9219 | 98.2063 | 96.2289 | 96.9645 |
| Read error rate/% | 0.253032 | 0.253032 | 0.229964 | 0.229964 | 0.238092 | 0.238092 |
| 数据库 Database | *版本或公开时间 Version or publication time |
|---|---|
| Gene Ontology(GO) | Releases_2017-09-08 |
| Kyoto Encyclopedia of Genes and Genomes,KEGG | v_81 |
| Cluster of Orthologous Groups of proteins,COG | Releases_2014-11-10 |
| Swiss-Pro | Release_2017-07 |
| Trembl | Release_2017-09 |
| NR | Release_2017-10-10 |
| EggNOG | v_4.5 |
| Antibiotic Resistance Genes Database,ARDB | v_1.1 |
| Pathogen Host Interactions,PHI | v_4.3 |
| Fungal Cytochrome P450 Database | v_1.1 |
| Carbohydrate-Active enzymes Database,CAZy | Release_2017-09 |
| Virulence Factor Database,VFDB | Release_2017-09 |
| Type III Secretion System Effector Proteins,T3SS | v_1.0 |
| TransportDB | v_2.0 |
表4 用于基因组注释的蛋白质数据库
Table 4 Protein database for genome annotation
| 数据库 Database | *版本或公开时间 Version or publication time |
|---|---|
| Gene Ontology(GO) | Releases_2017-09-08 |
| Kyoto Encyclopedia of Genes and Genomes,KEGG | v_81 |
| Cluster of Orthologous Groups of proteins,COG | Releases_2014-11-10 |
| Swiss-Pro | Release_2017-07 |
| Trembl | Release_2017-09 |
| NR | Release_2017-10-10 |
| EggNOG | v_4.5 |
| Antibiotic Resistance Genes Database,ARDB | v_1.1 |
| Pathogen Host Interactions,PHI | v_4.3 |
| Fungal Cytochrome P450 Database | v_1.1 |
| Carbohydrate-Active enzymes Database,CAZy | Release_2017-09 |
| Virulence Factor Database,VFDB | Release_2017-09 |
| Type III Secretion System Effector Proteins,T3SS | v_1.0 |
| TransportDB | v_2.0 |
| 基因组Genome | 序列类型 Seq type | 数目 Total number | 组装总长度 Total length/bp | 序列N50长度 N50 length/bp | 间隙数 Gap number | 序列GC含量 GC content /% |
|---|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Scaffold | 71 | 56 763 274 | 1 200 238 | 0 | 56.14 |
| Contig | 71 | 56 763 274 | 1 200 238 | - | 56.14 | |
| 线粒体基因组Mitochondrion genome | Scaffold | 1 | 85 353 | 85 353 | 0 | 26.91 |
| Contig | 1 | 85 353 | 85 353 | - | 26.91 |
表5 De novo测序组装得到的‘紫芝S2’全基因组
Table 5 Whole genome of Ganoderma sp. strain Zizhi S2* assembled by De novo sequencing
| 基因组Genome | 序列类型 Seq type | 数目 Total number | 组装总长度 Total length/bp | 序列N50长度 N50 length/bp | 间隙数 Gap number | 序列GC含量 GC content /% |
|---|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Scaffold | 71 | 56 763 274 | 1 200 238 | 0 | 56.14 |
| Contig | 71 | 56 763 274 | 1 200 238 | - | 56.14 | |
| 线粒体基因组Mitochondrion genome | Scaffold | 1 | 85 353 | 85 353 | 0 | 26.91 |
| Contig | 1 | 85 353 | 85 353 | - | 26.91 |
| Assembly length/bp | Coverage(#) | Coverage rate/% | Depth(#) | Reads usage percent/% |
|---|---|---|---|---|
| 56 848 627 | 56 499 028 | 99.39 | 177 | 83.45 |
表6 基因组序列深度、覆盖情况
Table 6 Genome sequencing depth and coverage
| Assembly length/bp | Coverage(#) | Coverage rate/% | Depth(#) | Reads usage percent/% |
|---|---|---|---|---|
| 56 848 627 | 56 499 028 | 99.39 | 177 | 83.45 |
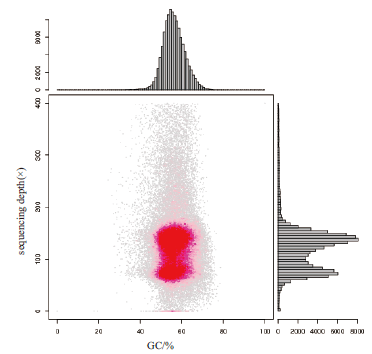
图1 GC含量与Depth关联分析统计图 横坐标是GC含量,纵坐标是平均深度
Fig.1 Statistical chart of correlation analysis between GC content and depth The abscissa is the GC content and the ordinate is the average depth
| 基因组 Genome | 类型 Type | 数目 Total number | 序列总长度 Total length/bp | 平均长度 Average length/bp | 占基因组总长的比例 Percentage in total length /% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Gene | 16 681 | 32 438 896 | 1 944.66 | 57.15 |
| Exons | 95 839 | 23 930 217 | 249.69 | 42.16 | |
| CDS | 16 681 | 23 930 217 | 1 434.58 | 42.16 | |
| Introne | 79 158 | 8 508 679 | 107.49 | 14.99 | |
| 线粒体基因组 Mitochondrion genome | Gene | 56 | 77 918 | 1 391.39 | 91.29 |
| Exons | 78 | 46 571 | 597.06 | 54.56 | |
| CDS | 56 | 46 571 | 831.62 | 54.56 | |
| Introne | 22 | 29 854 | 1 357.00 | 34.98 |
表7 基因组基因成分统计表
Table 7 Statistics of gene composition of the genome
| 基因组 Genome | 类型 Type | 数目 Total number | 序列总长度 Total length/bp | 平均长度 Average length/bp | 占基因组总长的比例 Percentage in total length /% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Gene | 16 681 | 32 438 896 | 1 944.66 | 57.15 |
| Exons | 95 839 | 23 930 217 | 249.69 | 42.16 | |
| CDS | 16 681 | 23 930 217 | 1 434.58 | 42.16 | |
| Introne | 79 158 | 8 508 679 | 107.49 | 14.99 | |
| 线粒体基因组 Mitochondrion genome | Gene | 56 | 77 918 | 1 391.39 | 91.29 |
| Exons | 78 | 46 571 | 597.06 | 54.56 | |
| CDS | 56 | 46 571 | 831.62 | 54.56 | |
| Introne | 22 | 29 854 | 1 357.00 | 34.98 |
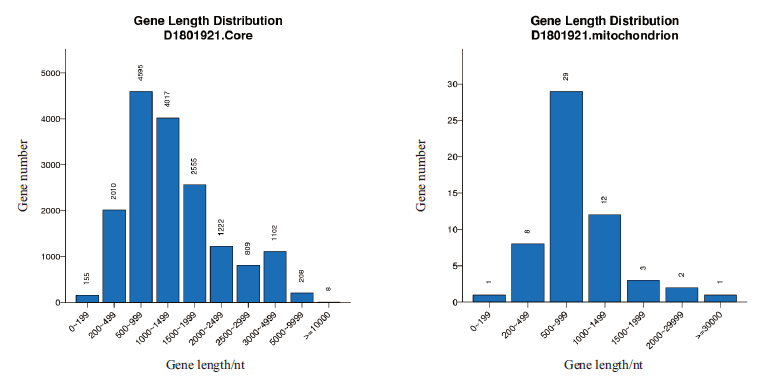
图2 核基因组与线粒体基因组中基因长度分布图 横坐标为基因长度,纵坐标为基因长度对应的基因数目
Fig. 2 Gene length distribution in nuclear genome and mitochondrial genome The abscissa is gene length,and the ordinate is the number of genes corresponding to the gene length
| Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number |
|---|---|---|---|---|---|---|---|---|
| Scaffold_1 | 4469875 | 1393 | Scaffold_31 | 666660 | 203 | Scaffold_53 | 207498 | 52 |
| Scaffold_10 | 1494552 | 424 | Scaffold_32 | 647059 | 167 | Scaffold_54 | 183478 | 51 |
| Scaffold_11 | 1421539 | 426 | Scaffold_33 | 627493 | 161 | Scaffold_55 | 176875 | 41 |
| Scaffold_12 | 1200238 | 334 | Scaffold_34 | 621976 | 186 | Scaffold_56 | 174271 | 58 |
| Scaffold_13 | 1081736 | 337 | Scaffold_35 | 614529 | 155 | Scaffold_57 | 167193 | 47 |
| Scaffold_14 | 1072049 | 347 | Scaffold_36 | 532656 | 153 | Scaffold_58 | 161880 | 47 |
| Scaffold_15 | 1050642 | 314 | Scaffold_37 | 511480 | 126 | Scaffold_59 | 161526 | 43 |
| Scaffold_16 | 1046259 | 337 | Scaffold_38 | 506477 | 122 | Scaffold_6 | 2277588 | 668 |
| Scaffold_17 | 1019206 | 303 | Scaffold_39 | 493120 | 139 | Scaffold_60 | 146901 | 39 |
| Scaffold_18 | 950046 | 247 | Scaffold_4 | 3153885 | 1014 | Scaffold_61 | 115424 | 31 |
| Scaffold_19 | 926904 | 298 | Scaffold_40 | 428051 | 134 | Scaffold_62 | 114060 | 27 |
| Scaffold_2 | 3484778 | 1149 | Scaffold_41 | 415410 | 111 | Scaffold_63 | 103079 | 35 |
| Scaffold_20 | 895893 | 264 | Scaffold_42 | 375402 | 111 | Scaffold_64 | 95381 | 24 |
| Scaffold_21 | 879786 | 257 | Scaffold_43 | 333682 | 104 | Scaffold_65 | 86665 | 14 |
| Scaffold_22 | 879764 | 234 | Scaffold_44 | 331534 | 85 | Scaffold_66 | 83587 | 23 |
| Scaffold_23 | 848793 | 236 | Scaffold_45 | 319851 | 90 | Scaffold_67 | 78814 | 27 |
| Scaffold_24 | 842024 | 258 | Scaffold_46 | 311630 | 87 | Scaffold_68 | 66934 | 13 |
| Scaffold_25 | 790967 | 201 | Scaffold_47 | 282820 | 90 | Scaffold_69 | 59421 | 15 |
| Scaffold_26 | 790090 | 218 | Scaffold_48 | 257692 | 74 | Scaffold_7 | 2147372 | 600 |
| Scaffold_27 | 747333 | 216 | Scaffold_49 | 248,648 | 51 | Scaffold_70 | 58166 | 16 |
| Scaffold_28 | 714016 | 198 | Scaffold_5 | 2522070 | 731 | Scaffold_71 | 33177 | 15 |
| Scaffold_29 | 692498 | 211 | Scaffold_50 | 232229 | 58 | Scaffold_72 | 85353 | 56 |
| Scaffold_3 | 3315994 | 946 | Scaffold_51 | 226670 | 86 | Scaffold_8 | 2100477 | 654 |
| Scaffold_30 | 691292 | 185 | Scaffold_52 | 212908 | 70 | Scaffold_9 | 1783301 | 500 |
表8 Scaffold 各条长度及其分布的基因数目统计
Table 8 Statistics on the total sequence length and genes’ number on each scaffold
| Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number |
|---|---|---|---|---|---|---|---|---|
| Scaffold_1 | 4469875 | 1393 | Scaffold_31 | 666660 | 203 | Scaffold_53 | 207498 | 52 |
| Scaffold_10 | 1494552 | 424 | Scaffold_32 | 647059 | 167 | Scaffold_54 | 183478 | 51 |
| Scaffold_11 | 1421539 | 426 | Scaffold_33 | 627493 | 161 | Scaffold_55 | 176875 | 41 |
| Scaffold_12 | 1200238 | 334 | Scaffold_34 | 621976 | 186 | Scaffold_56 | 174271 | 58 |
| Scaffold_13 | 1081736 | 337 | Scaffold_35 | 614529 | 155 | Scaffold_57 | 167193 | 47 |
| Scaffold_14 | 1072049 | 347 | Scaffold_36 | 532656 | 153 | Scaffold_58 | 161880 | 47 |
| Scaffold_15 | 1050642 | 314 | Scaffold_37 | 511480 | 126 | Scaffold_59 | 161526 | 43 |
| Scaffold_16 | 1046259 | 337 | Scaffold_38 | 506477 | 122 | Scaffold_6 | 2277588 | 668 |
| Scaffold_17 | 1019206 | 303 | Scaffold_39 | 493120 | 139 | Scaffold_60 | 146901 | 39 |
| Scaffold_18 | 950046 | 247 | Scaffold_4 | 3153885 | 1014 | Scaffold_61 | 115424 | 31 |
| Scaffold_19 | 926904 | 298 | Scaffold_40 | 428051 | 134 | Scaffold_62 | 114060 | 27 |
| Scaffold_2 | 3484778 | 1149 | Scaffold_41 | 415410 | 111 | Scaffold_63 | 103079 | 35 |
| Scaffold_20 | 895893 | 264 | Scaffold_42 | 375402 | 111 | Scaffold_64 | 95381 | 24 |
| Scaffold_21 | 879786 | 257 | Scaffold_43 | 333682 | 104 | Scaffold_65 | 86665 | 14 |
| Scaffold_22 | 879764 | 234 | Scaffold_44 | 331534 | 85 | Scaffold_66 | 83587 | 23 |
| Scaffold_23 | 848793 | 236 | Scaffold_45 | 319851 | 90 | Scaffold_67 | 78814 | 27 |
| Scaffold_24 | 842024 | 258 | Scaffold_46 | 311630 | 87 | Scaffold_68 | 66934 | 13 |
| Scaffold_25 | 790967 | 201 | Scaffold_47 | 282820 | 90 | Scaffold_69 | 59421 | 15 |
| Scaffold_26 | 790090 | 218 | Scaffold_48 | 257692 | 74 | Scaffold_7 | 2147372 | 600 |
| Scaffold_27 | 747333 | 216 | Scaffold_49 | 248,648 | 51 | Scaffold_70 | 58166 | 16 |
| Scaffold_28 | 714016 | 198 | Scaffold_5 | 2522070 | 731 | Scaffold_71 | 33177 | 15 |
| Scaffold_29 | 692498 | 211 | Scaffold_50 | 232229 | 58 | Scaffold_72 | 85353 | 56 |
| Scaffold_3 | 3315994 | 946 | Scaffold_51 | 226670 | 86 | Scaffold_8 | 2100477 | 654 |
| Scaffold_30 | 691292 | 185 | Scaffold_52 | 212908 | 70 | Scaffold_9 | 1783301 | 500 |
| 基因组 Genome | 类型 Type | 拷贝数目 Number of copies | 平均长度 Average length/bp | 总长度 Total length/bp | 占基因组百分比 Percentage in genome/% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | tRNA | 226 | 81.04 | 18 316 | 0.0323 |
| rRNA(by De novo prediction)* | 12 | 2 883.08 | 34 597 | 0.0609 | |
| sRNA | 64 | 62.23 | 3 983 | 0.007 | |
| snRNA | 29 | 102.79 | 2 981 | 0.0053 | |
| miRNA | 74 | 59.91 | 4 434 | 0.0078 | |
| 线粒体基因组 Mitochondrion genome | tRNA | 20 | 72.9 | 1 458 | 1.7082 |
| rRNA(by De novo prediction)** | 0 | 0 | 0 | 0 | |
| sRNA | 1 | 90 | 90 | 0.1054 | |
| snRNA | 1 | 38 | 38 | 0.0445 | |
| miRNA | 1 | 88 | 88 | 0.1031 |
表9 非编码RNA统计
Table 9 Statistics of non-coding RNA
| 基因组 Genome | 类型 Type | 拷贝数目 Number of copies | 平均长度 Average length/bp | 总长度 Total length/bp | 占基因组百分比 Percentage in genome/% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | tRNA | 226 | 81.04 | 18 316 | 0.0323 |
| rRNA(by De novo prediction)* | 12 | 2 883.08 | 34 597 | 0.0609 | |
| sRNA | 64 | 62.23 | 3 983 | 0.007 | |
| snRNA | 29 | 102.79 | 2 981 | 0.0053 | |
| miRNA | 74 | 59.91 | 4 434 | 0.0078 | |
| 线粒体基因组 Mitochondrion genome | tRNA | 20 | 72.9 | 1 458 | 1.7082 |
| rRNA(by De novo prediction)** | 0 | 0 | 0 | 0 | |
| sRNA | 1 | 90 | 90 | 0.1054 | |
| snRNA | 1 | 38 | 38 | 0.0445 | |
| miRNA | 1 | 88 | 88 | 0.1031 |
| Genome | Method | Repeat size/bp | Percentage in genome/% |
|---|---|---|---|
| 核基因组 Nuclear genome | Repbase | 1 307 464 | 2.3034 |
| ProMask | 1 973 669 | 3.477 | |
| De novo | 6 991 719 | 12.3173 | |
| TRF | 326 000 | 0.5743 | |
| Total | 7 954 327 | 14.0132 | |
| 线粒体基因组 Mitochondrion genome | Repbase | 128 | 0.15 |
| ProMask | 1 005 | 1.1775 | |
| De novo | 0 | ||
| TRF | 320 | 0.3749 | |
| Total | 1 453 | 1.7023 |
表10 重复序列统计
Table 10 Statistics of repeat sequences in the genome
| Genome | Method | Repeat size/bp | Percentage in genome/% |
|---|---|---|---|
| 核基因组 Nuclear genome | Repbase | 1 307 464 | 2.3034 |
| ProMask | 1 973 669 | 3.477 | |
| De novo | 6 991 719 | 12.3173 | |
| TRF | 326 000 | 0.5743 | |
| Total | 7 954 327 | 14.0132 | |
| 线粒体基因组 Mitochondrion genome | Repbase | 128 | 0.15 |
| ProMask | 1 005 | 1.1775 | |
| De novo | 0 | ||
| TRF | 320 | 0.3749 | |
| Total | 1 453 | 1.7023 |
| Genome | Method | Type | DNA | LINE | LTR | SINE | Other | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|
| 核基因组Nuclear genome | Repbase TEs | Length(bp) | 95 474 | 66 559 | 1 146 867 | 2 711 | 165 | 1 360 | 1 307 464 |
| % in Genome | 0.1682 | 0.1173 | 2.0204 | 0.0048 | 0.0003 | 0.0024 | 2.3034 | ||
| ProteinMask TEs | Length(bp) | 316 367 | 100 469 | 1 558 079 | 0 | 0 | 0 | 1 973 669 | |
| % in Genome | 0.5573 | 0.177 | 2.7449 | 0 | 0 | 0 | 3.477 | ||
| De novo TEs | Length(bp) | 594 332 | 431 488 | 2 449 588 | 4 081 | 0 | 3 592 915 | 6 991 719 | |
| % in Genome | 1.047 | 0.7602 | 4.3154 | 0.0072 | 0 | 6.3296 | 12.3173 | ||
| Combined TEs | Length(bp) | 924 792 | 515 460 | 2 849 776 | 5 828 | 165 | 3 594 275 | 7 718 181 | |
| % in Genome | 1.6292 | 0.9081 | 5.0205 | 0.0103 | 0.0003 | 6.332 | 13.5971 | ||
| 线粒体基因组 Mitochondrion genome | Repbase TEs | Length(bp) | 0 | 0 | 128 | 0 | 0 | 0 | 128 |
| % in Genome | 0 | 0 | 0.15 | 0 | 0 | 0 | 0.15 | ||
| ProteinMask TEs | Length(bp) | 0 | 1 005 | 0 | 0 | 0 | 0 | 1 005 | |
| % in Genome | 0 | 1.1775 | 0 | 0 | 0 | 0 | 1.1775 | ||
| De novo TEs | Length(bp) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| % in Genome | - | - | - | - | - | - | - | ||
| Combined TEs | Length(bp) | 0 | 1 005 | 128 | 0 | 0 | 0 | 1 133 | |
| % in Genome | 0 | 1.1775 | 0.15 | 0 | 0 | 0 | 1.3274 |
表11 转座子分类统计表
Table 11 Statistics of transposable factors
| Genome | Method | Type | DNA | LINE | LTR | SINE | Other | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|
| 核基因组Nuclear genome | Repbase TEs | Length(bp) | 95 474 | 66 559 | 1 146 867 | 2 711 | 165 | 1 360 | 1 307 464 |
| % in Genome | 0.1682 | 0.1173 | 2.0204 | 0.0048 | 0.0003 | 0.0024 | 2.3034 | ||
| ProteinMask TEs | Length(bp) | 316 367 | 100 469 | 1 558 079 | 0 | 0 | 0 | 1 973 669 | |
| % in Genome | 0.5573 | 0.177 | 2.7449 | 0 | 0 | 0 | 3.477 | ||
| De novo TEs | Length(bp) | 594 332 | 431 488 | 2 449 588 | 4 081 | 0 | 3 592 915 | 6 991 719 | |
| % in Genome | 1.047 | 0.7602 | 4.3154 | 0.0072 | 0 | 6.3296 | 12.3173 | ||
| Combined TEs | Length(bp) | 924 792 | 515 460 | 2 849 776 | 5 828 | 165 | 3 594 275 | 7 718 181 | |
| % in Genome | 1.6292 | 0.9081 | 5.0205 | 0.0103 | 0.0003 | 6.332 | 13.5971 | ||
| 线粒体基因组 Mitochondrion genome | Repbase TEs | Length(bp) | 0 | 0 | 128 | 0 | 0 | 0 | 128 |
| % in Genome | 0 | 0 | 0.15 | 0 | 0 | 0 | 0.15 | ||
| ProteinMask TEs | Length(bp) | 0 | 1 005 | 0 | 0 | 0 | 0 | 1 005 | |
| % in Genome | 0 | 1.1775 | 0 | 0 | 0 | 0 | 1.1775 | ||
| De novo TEs | Length(bp) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| % in Genome | - | - | - | - | - | - | - | ||
| Combined TEs | Length(bp) | 0 | 1 005 | 128 | 0 | 0 | 0 | 1 133 | |
| % in Genome | 0 | 1.1775 | 0.15 | 0 | 0 | 0 | 1.3274 |
| 数据库 | Total | P450 | TF | CAZY | PHI | IPR |
|---|---|---|---|---|---|---|
| Nuclear genome | 16 681 | 1 261(7.55%) | 588(3.52%) | 278(1.66%) | 570(3.41%) | 10 296(61.72%) |
| Mitochondrial genome | 56 | 0(0%) | 0(0%) | 0(0%) | 1(1.78%) | 47(83.92%) |
| 数据库 | SWISSPROT | COG | GO | KEGG | NR | Over All |
| Nuclear genome | 2 708(16.23%) | 1 244(7.45%) | 7 373(44.19%) | 4 346(26.05%) | 12 161(72.9%) | 13 161(78.89%) |
| Mitochondrial genome | 24(42.85%) | 10(17.85%) | 44(78.57%) | 16(28.57%) | 46(82.14%) | 51(91.07%) |
表12 基因集注释结果统计
Table 12 Statistics of gene set annotation results
| 数据库 | Total | P450 | TF | CAZY | PHI | IPR |
|---|---|---|---|---|---|---|
| Nuclear genome | 16 681 | 1 261(7.55%) | 588(3.52%) | 278(1.66%) | 570(3.41%) | 10 296(61.72%) |
| Mitochondrial genome | 56 | 0(0%) | 0(0%) | 0(0%) | 1(1.78%) | 47(83.92%) |
| 数据库 | SWISSPROT | COG | GO | KEGG | NR | Over All |
| Nuclear genome | 2 708(16.23%) | 1 244(7.45%) | 7 373(44.19%) | 4 346(26.05%) | 12 161(72.9%) | 13 161(78.89%) |
| Mitochondrial genome | 24(42.85%) | 10(17.85%) | 44(78.57%) | 16(28.57%) | 46(82.14%) | 51(91.07%) |
| Species and strain name | G. sp. Zizhi S2 | G. lucidum G.260125-1 | G. sinense ZZ0214-1 |
|---|---|---|---|
| Chromosomes | NA* | 13 | 12 |
| Number of scaffolds | 71 | 82 | 69 |
| Length of genome assembly(bp) | 56 763 274 | 43 292 570 | 48 955 697 |
| GC content(%) | 56.14 | 56.16 | 56.17 |
| Number of protein-coding genes | 16 681 | 16 495 | 15 688 |
| Average gene length(bp) | 1 944.66 | 1 569.02 | 1 668.82 |
| GC content of protein-coding genes(%) | 59.62 | 59.33 | 59.53 |
| Average number of exons per gene | 5.74 | 4.58 | 5.34 |
| Average exon size(bp) | 249.69 | 259 | 245.67 |
| Average coding sequence size(bp) | 1 434.57 | 1 188.79 | 1 313.03 |
| Average intron size(bp) | 107.24 | 94.12 | 81.71 |
| Average size of intergenic regions(bp) | 1 452.02 | 1 050.52 | 1 487.12 |
| Repeat sequences(%) | 14.01 | 8.15 | 8.53 |
| Sequencing platform | PacBio Sequel & Illumina HiSeq | Roche 450 & Illumina | Roche 454 & Illumina HiSeq |
表13 三个灵芝核基因组信息比较
Table 13 Information comparison of 3 nuclear genomes of Ganoderma spp.
| Species and strain name | G. sp. Zizhi S2 | G. lucidum G.260125-1 | G. sinense ZZ0214-1 |
|---|---|---|---|
| Chromosomes | NA* | 13 | 12 |
| Number of scaffolds | 71 | 82 | 69 |
| Length of genome assembly(bp) | 56 763 274 | 43 292 570 | 48 955 697 |
| GC content(%) | 56.14 | 56.16 | 56.17 |
| Number of protein-coding genes | 16 681 | 16 495 | 15 688 |
| Average gene length(bp) | 1 944.66 | 1 569.02 | 1 668.82 |
| GC content of protein-coding genes(%) | 59.62 | 59.33 | 59.53 |
| Average number of exons per gene | 5.74 | 4.58 | 5.34 |
| Average exon size(bp) | 249.69 | 259 | 245.67 |
| Average coding sequence size(bp) | 1 434.57 | 1 188.79 | 1 313.03 |
| Average intron size(bp) | 107.24 | 94.12 | 81.71 |
| Average size of intergenic regions(bp) | 1 452.02 | 1 050.52 | 1 487.12 |
| Repeat sequences(%) | 14.01 | 8.15 | 8.53 |
| Sequencing platform | PacBio Sequel & Illumina HiSeq | Roche 450 & Illumina | Roche 454 & Illumina HiSeq |
| [1] | 钟礼义. 紫芝新品种武芝2号区域试验[J]. 中国食用菌, 2013, 32(6): 25-27. |
| Zhong LY. The regional test on new varieties of Ganoderma[J]. Edible Fungi China, 2013, 32(6): 25-27. | |
| [2] | 钟礼义, 陈体强, 刘新锐, 等. 鉴别紫芝菌株的PCR引物筛选及其序列比对验证[J]. 福建农业学报, 2020, 35(7): 725-730. |
| Zhong LY, Chen TQ, Liu XR, et al. Selection and sequence alignment of PCR primers for identifying Zizhi strain[J]. Fujian J Agric Sci, 2020, 35(7): 725-730. | |
| [3] | 陈体强, 吴锦忠, 钟礼义, 等. 福建野生紫芝资源的开发利用Ⅰ. 硬孔灵芝[J]. 福建农业大学学报:自然科学版, 2006, 35(3): 324-328. |
| Chen TQ, Wu JZ, Zhong LY, et al. Exploitation and utilization of wild Zhizi resource in Fujian Ⅰ. Ganoderma duropora(Sect. Phaeonema)[J]. J Fujian Agric For Univ:Nat Sci Edn, 2006, 35(3): 324-328. | |
| [4] | Hapuarachchi KK, Karunarathna SC, McKenzie EHC, et al. High phenotypic plasticity of Ganoderma sinense(Ganodermataceae, Polyporales)in China[J]. Asian J Mycol, 2019, 2(1): 1-47. |
| [5] | Wang DM, Zhang XQ, Yao YJ. Type studies of some Ganoderma species from China[J]. Mycotaxon, 2005, 93: 61-70. |
| [6] | 王冬梅. 中国灵芝属系统发育研究[D]. 北京:中国科学院微生物研究所, 2005. |
| Wang DM. Phylogeny of Ganoderma in China[D]. Beijing:Institute of Microbiology, Chinese Academy of Sciences, 2005. | |
| [7] | 曹云. 中国灵芝属的系统学研究[D]. 北京:中国科学院大学, 2013. |
| Cao Y. Systematic study of Ganoderma in China[D]. Beijing:University of Chinese Academy of Sciences, 2013. | |
| [8] | 王新存. 灵芝科系统发育研究[D]. 北京:中国科学院大学, 2012. |
| Wang XC. Phylogenetic studies of Ganodermataceae. Beijing:University of Chinese Academy of Sciences, 2012. | |
| [9] | 邢佳慧. 灵芝属的物种多样性、分类与系统发育研究[D]. 北京:北京林业大学, 2019. |
| Xing JH. Species diversity, taxonomy and phylogeny of Ganoderma[D]. Beijing:Beijing Forestry University, 2019. | |
| [10] | 陈体强, 吴锦忠, 李晔, 等. 福建野生紫芝资源开发利用Ⅱ. ‘闽紫96’(中国灵芝)[J]. 菌物研究, 2006, 4(4): 27-32. |
| Chen TQ, Wu JZ, Li Y, et al. Exploitation and utilization of wild Zhizi resources(Sect. Phaeonema of Ganoderma)in Fujian(Ⅱ). G. sinense ‘minzi 96’[J]. J Fungal Res, 2006, 4(4): 27-32. | |
| [11] |
Kim KE, Peluso P, Babayan P, et al. Long-read, whole-genome shotgun sequence data for five model organisms[J]. Sci Data, 2014, 1: 140045.
doi: 10.1038/sdata.2014.45 URL |
| [12] | Faino L, Seidl MF, Datema E, et al. Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome[J]. mBio, 2015, 6(4): e00936-15. |
| [13] |
Sit CS, Ruzzini AC, Van Arnam EB, et al. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants[J]. PNAS, 2015, 112(43): 13150-13154.
doi: 10.1073/pnas.1515348112 URL |
| [14] | Tsuji M, Kudoh S, Hoshino T. Draft genome sequence of cryophilic basidiomycetous yeast Mrakia blollopis SK-4, isolated from an algal mat of Naga-ike Lake in the Skarvsnes ice-free area, East Antarctica[J]. Genome Announc, 2015, 3(1): e01454-14. |
| [15] |
Badouin H, Hood ME, Gouzy J, et al. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae[J]. Genetics, 2015, 200(4): 1275-1284.
doi: 10.1534/genetics.115.177709 pmid: 26044594 |
| [16] |
Fei X, Zhao MW, Li YX. Cloning and sequence analysis of a glyceraldehyde-3-phosphate dehydrogenase gene from Ganoderma lucidum[J]. J Microbiol, 2006, 44(5): 515-522.
pmid: 17082745 |
| [17] | 张妍, 黄晨阳, 高巍. 食用菌分子育种研究进展[J]. 菌物研究, 2019, 17(4): 229-239. |
| Zhang Y, Huang CY, Gao W. Research advances on molecular mushroom breeding[J]. J Fungal Res, 2019, 17(4): 229-239. | |
| [18] | 左斌, 王海斌, 葛俊, 等. 真菌漆酶基因研究进展[J]. 微生物学免疫学进展, 2009, 37(1): 72-76. |
| Zuo B, Wang HB, Ge J, et al. Research progress in the gene of fungal laccase[J]. Prog Microbiol Immunol, 2009, 37(1): 72-76. | |
| [19] |
Sun SJ, Liu JZ, Hu KH, et al. The level of secreted laccase activity in the edible fungi and their growing cycles are closely related[J]. Curr Microbiol, 2011, 62(3): 871-875.
doi: 10.1007/s00284-010-9794-z pmid: 21046396 |
| [20] | 陈体强, 吴建国, 毛方华, 等. 硬孔灵芝和赤芝孢子油的脂肪酸、角鲨烯及麦角甾醇类的比较[J]. 食用菌学报, 2018, 25(1): 47-52. |
| Chen TQ, Wu JG, Mao FH, et al. Comparison on fatty acid profile, squalene and ergosterols between spore oils extracted from Ganoderma duropora and G. lucidum[J]. Acta Edulis Fungi, 2018, 25(1): 47-52. | |
| [21] |
Chen TQ, Wu YB, Wu JG, et al. Fatty acids, essential oils, and squalene in the spore lipids of Ganoderma lucidum by GC-MS and GC-FID[J]. Chem Nat Compd, 2013, 49(1): 143-144.
doi: 10.1007/s10600-013-0536-x URL |
| [22] |
Lian CL, Wu YQ, Chen TQ, et al. Identification of new trace triterpenoids from the fungus Ganoderma duripora[J]. Phytochem Lett, 2017, 21: 237-239.
doi: 10.1016/j.phytol.2017.07.005 URL |
| [23] |
Chen TQ, Wu JG, Wu YB, et al. Supercritical fluid CO2 extraction, simultaneous determination of total sterols in the spore lipids of Ganoderma lucidum by GC-MS/SIM methods[J]. Chem Nat Compd, 2012, 48(4): 657-658.
doi: 10.1007/s10600-012-0338-6 URL |
| [24] | 赵芬, 李晔, 刘超, 等. 硬孔灵芝的化学成分研究[J]. 菌物学报, 2009, 28(3): 407-409. |
| Zhao F, Li Y, Liu C, et al. Chemical components from the fruiting bodies of Ganoderma duropora[J]. Mycosystema, 2009, 28(3): 407-409. | |
| [25] |
Lian CL, Wang CF, Xiao Q, et al. The triterpenes and steroids from the fruiting body Ganoderma duripora[J]. Biochem Syst Ecol, 2017, 73: 50-53.
doi: 10.1016/j.bse.2017.06.005 URL |
| [26] | 方星, 师亮, 徐颖洁, 等. 灵芝甾醇14α-脱甲基酶基因的克隆及超量表达对三萜合成的影响[J]. 菌物学报, 2011, 30(2): 242-248. |
| Fang X, Shi L, Xu YJ, et al. Cloning of a sterol 14α-demethylase gene and the effects of over-expression of the gene on biological synjournal of triterpenes in Ganoderma lucidum[J]. Mycosystema, 2011, 30(2): 242-248. | |
| [27] | Grigoriev IV, Dullen D, Goodwin SB, et al. Fueling the future with fungal genomics[J]. Mycology, 2011, 2(3): 192-209. |
| [28] |
Grigoriev IV, Nikitin R, Haridas S, et al. MycoCosm portal:gearing up for 1000 fungal genomes[J]. Nucleic Acids Res, 2014, 42(database issue): D699-D704.
doi: 10.1093/nar/gkt1183 URL |
| [29] | 刘淑艳, 李广, 李玉. 菌物基因组测定研究进展[J]. 吉林农业大学学报, 2014, 36(1): 1-9, 16. |
| Liu SY, Li G, Li Y. Advance of fungal genomes sequencing researches[J]. J Jilin Agric Univ, 2014, 36(1): 1-9, 16. | |
| [30] | 凌志琳, 赵瑞琳. 基因组学在食药用菌栽培育种中的研究进展[J]. 食用菌学报, 2018, 25(1): 93-106. |
| Ling ZL, Zhao RL. Advances of genomics-assisted cultivation and breeding of edible and medicinal mushrooms[J]. Acta Edulis Fungi, 2018, 25(1): 93-106. | |
| [31] |
Chen SL, Xu J, Liu C, et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum[J]. Nat Commun, 2012, 3: 913.
doi: 10.1038/ncomms1923 URL |
| [32] |
Zhu YJ, Xu J, Sun C, et al. Chromosome-level genome map provides insights into diverse defense mechanisms in the medicinal fungus Ganoderma sinense[J]. Sci Rep, 2015, 5: 11087.
doi: 10.1038/srep11087 URL |
| [33] |
Benveniste P. Biosynjournal and accumulation of sterols[J]. Annu Rev Plant Biol, 2004, 55: 429-457.
pmid: 15377227 |
| [1] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [2] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [3] | 郭少华, 毛会丽, 刘征权, 付美媛, 赵平原, 马文博, 李旭东, 关建义. 一株鱼源致病性嗜水气单胞菌XDMG的全基因组测序及比较基因组分析[J]. 生物技术通报, 2023, 39(8): 291-306. |
| [4] | 张志霞, 李天培, 曾虹, 朱稀贤, 杨天雄, 马斯楠, 黄磊. 冰冷杆菌PG-2的基因组测序及生物信息学分析[J]. 生物技术通报, 2023, 39(3): 290-300. |
| [5] | 和梦颖, 刘文彬, 林震鸣, 黎尔彤, 汪洁, 金小宝. 一株抗革兰阳性菌的戈登氏菌WA4-43全基因组测序与分析[J]. 生物技术通报, 2023, 39(2): 232-242. |
| [6] | 张傲洁, 李青云, 宋文红, 颜少慧, 唐爱星, 刘幽燕. 基于苯酚降解的粪产碱杆菌Alcaligenes faecalis JF101的全基因组分析[J]. 生物技术通报, 2023, 39(10): 292-303. |
| [7] | 王帅, 吕鸿睿, 张昊, 吴占文, 肖翠红, 孙冬梅. 解磷菌PSB-R全基因组测序鉴定及其解磷特性分析[J]. 生物技术通报, 2023, 39(1): 274-283. |
| [8] | 张泽颖, 范清锋, 邓云峰, 韦廷舟, 周正富, 周建, 王劲, 江世杰. 一株高产脂肪酶菌株WCO-9全基因组测序及比较基因组分析[J]. 生物技术通报, 2022, 38(10): 216-225. |
| [9] | 薛清, 杜虹锐, 薛会英, 王译浩, 王暄, 李红梅. 苜蓿滑刃线虫线粒体基因组及其系统发育研究[J]. 生物技术通报, 2021, 37(7): 98-106. |
| [10] | 郭鹤宝, 王星, 何山文, 张晓霞. 表型特征结合基因组分析鉴定不同菌落形态Bacillus velezensis ACCC 19742[J]. 生物技术通报, 2020, 36(2): 142-148. |
| [11] | 李晓凯 ,王贵 ,乔贤 ,范一星 ,张磊 ,马宇浩 ,聂瑞雪 ,王瑞军 ,何利兵 ,苏蕊. 全基因组测序在重要家畜上的研究进展[J]. 生物技术通报, 2018, 34(6): 11-21. |
| [12] | 王兴文, 王加启, 赵圣国, 李发弟, 卜登攀. 未培养技术在瘤胃产甲烷菌群研究中的应用[J]. 生物技术通报, 2014, 0(6): 67-74. |
| [13] | 索志立;. 利用DNA ISSR分子标记技术对芍药属植物栽培品种的分类鉴定方法[J]. , 2008, 0(S1): 109-112. |
| [14] | Margarita Escaler;汪开治. 亚洲农业生物技术发展现状和前景(下)[J]. , 2005, 0(04): 75-78. |
| [15] | 孙国凤;. 利用细胞质雄性不育系和恢复系育成番茄栽培种[J]. , 1997, 0(01): 24-25. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||