








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 136-147.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0533
赖瑞联( ), 冯新, 高敏霞, 路喻丹, 刘晓驰, 吴如健, 陈义挺(
), 冯新, 高敏霞, 路喻丹, 刘晓驰, 吴如健, 陈义挺( )
)
收稿日期:2022-04-29
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
陈义挺,男,研究员,研究方向:果树生物技术与遗传资源;E-mail: chyiting@163.com作者简介:赖瑞联,男,助理研究员,研究方向:果树生物技术与遗传资源;E-mail: lairl0618@163.com
基金资助:
LAI Rui-lian( ), FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting(
), FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting( )
)
Received:2022-04-29
Published:2023-04-26
Online:2023-05-16
摘要:
过氧化氢酶(Catalase,CAT)是植物主要的抗氧化酶之一。为揭示猕猴桃CAT(AcCAT)基因家族序列特征和潜在功能,对其进行了全基因组鉴定和表达分析。对AcCAT基因家族进行全基因组鉴定和生物信息学分析,并研究其在不同器官以及果实贮藏过程中的表达变化。从猕猴桃基因组中鉴定出9个AcCAT基因并根据其在染色体上的位置分别命名为AcCAT1-AcCAT9,不同成员之间蛋白理化性质、基因结构、保守基序、启动子作用元件等存在较高相似性。AcCAT基因不均匀地分布在猕猴桃的4条染色体上,其中的5个AcCAT基因形成2个串联基因簇,6个AcCAT基因发生片段复制。基于psRNAtarget分析,AcCAT基因可能主要受miR166家族调控。系统进化分析结果显示,来源于猕猴桃、茶树、棉花和水稻的23个CAT蛋白分为3个类群,其归类并未完全按物种划分。在不同猕猴桃器官中,AcCAT3主要在根中表达,AcCAT1和AcCAT4主要在花中表达,AcCAT2、AcCAT5、AcCAT6、AcCAT8和AcCAT9主要在叶中表达,AcCAT7则同时在根和花中高水平表达;在猕猴桃果实贮藏过程中,AcCAT5和AcCAT6表达水平逐渐降低,AcCAT1和AcCAT2主要在贮藏前期表达,而AcCAT3、AcCAT4和AcCAT8主要在贮藏后期表达。此外,miR166家族成员在果实贮藏过程中的表达量逐步升高。AcCAT基因在进化过程中存在保守性,其在猕猴桃生长发育和果实贮藏软化过程中发挥调控作用。
赖瑞联, 冯新, 高敏霞, 路喻丹, 刘晓驰, 吴如健, 陈义挺. 猕猴桃过氧化氢酶基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2023, 39(4): 136-147.
LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit[J]. Biotechnology Bulletin, 2023, 39(4): 136-147.
| 蛋白名称 Protein name | 氨基酸 Amino acids | 分子量 Molecular weight/kD | 等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲水性系数 Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| AcCAT1 | 450 | 52.16 | 6.85 | 39.98 | 69.93 | -0.585 |
| AcCAT2 | 757 | 87.31 | 7.24 | 48.49 | 72.60 | -0.631 |
| AcCAT3 | 1 285 | 148.21 | 5.89 | 42.86 | 79.04 | -0.420 |
| AcCAT4 | 459 | 53.40 | 7.82 | 38.03 | 76.06 | -0.497 |
| AcCAT5 | 501 | 58.01 | 6.39 | 44.37 | 71.40 | -0.537 |
| AcCAT6 | 526 | 60.67 | 6.53 | 41.73 | 70.06 | -0.535 |
| AcCAT7 | 302 | 35.33 | 6.09 | 44.08 | 73.61 | -0.542 |
| AcCAT8 | 490 | 57.08 | 6.88 | 38.26 | 69.63 | -0.536 |
| AcCAT9 | 91 | 10.34 | 5.90 | 37.97 | 63.19 | -0.676 |
表1 猕猴桃AcCAT蛋白的理化性质
Table 1 Physicochemical properties of AcCAT proteins in A. chinensis
| 蛋白名称 Protein name | 氨基酸 Amino acids | 分子量 Molecular weight/kD | 等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲水性系数 Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| AcCAT1 | 450 | 52.16 | 6.85 | 39.98 | 69.93 | -0.585 |
| AcCAT2 | 757 | 87.31 | 7.24 | 48.49 | 72.60 | -0.631 |
| AcCAT3 | 1 285 | 148.21 | 5.89 | 42.86 | 79.04 | -0.420 |
| AcCAT4 | 459 | 53.40 | 7.82 | 38.03 | 76.06 | -0.497 |
| AcCAT5 | 501 | 58.01 | 6.39 | 44.37 | 71.40 | -0.537 |
| AcCAT6 | 526 | 60.67 | 6.53 | 41.73 | 70.06 | -0.535 |
| AcCAT7 | 302 | 35.33 | 6.09 | 44.08 | 73.61 | -0.542 |
| AcCAT8 | 490 | 57.08 | 6.88 | 38.26 | 69.63 | -0.536 |
| AcCAT9 | 91 | 10.34 | 5.90 | 37.97 | 63.19 | -0.676 |

图4 猕猴桃AcCAT基因在染色体上的分布 ①为染色体基因密度,②为GC含量,③为GC偏移,④为未知碱基分布
Fig. 4 Distribution of AcCAT genes on the chromosomes of A. chinensis ①indicates chromosome gene density, ② indicates GC ratio, ③ indicates GC skew, and ④ indicates Nratio
| miRNA | 靶基因Target | 期望值Expectation | 序列比对Alignment | 抑制方式Inhibition |
|---|---|---|---|---|
| ach-miR166k-3p | AcCAT5 | 2.0 | miRNA 21 UCCCUAACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166k-3p | AcCAT6 | 2.0 | miRNA 21 UCCCUAACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a-3p | AcCAT6 | 3.0 | miRNA 21 CCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a-3p | AcCAT5 | 3.0 | miRNA 21 CCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166b | AcCAT6 | 3.0 | miRNA 21 AUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166b | AcCAT5 | 3.0 | miRNA 21 AUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166g-3p | AcCAT5 | 3.0 | miRNA 21 CUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166g-3p | AcCAT6 | 3.0 | miRNA 21 CUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166m | AcCAT5 | 3.0 | miRNA 21 UCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166m | AcCAT6 | 3.0 | miRNA 21 UCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166n | AcCAT5 | 3.0 | miRNA 21 UUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166n | AcCAT6 | 3.0 | miRNA 21 UUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a | AcCAT6 | 3.0 | miRNA 19 CCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1451 GGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a | AcCAT5 | 3.0 | miRNA 19 CCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1340 GGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166h-3p | AcCAT5 | 3.0 | miRNA 20 CCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1339 UGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166h-3p | AcCAT6 | 3.0 | miRNA 20 CCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1450 UGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
表2 靶向调控猕猴桃AcCAT基因的miRNA
Table 2 miRNAs targeting regulating AcCAT genes in A. chinensis
| miRNA | 靶基因Target | 期望值Expectation | 序列比对Alignment | 抑制方式Inhibition |
|---|---|---|---|---|
| ach-miR166k-3p | AcCAT5 | 2.0 | miRNA 21 UCCCUAACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166k-3p | AcCAT6 | 2.0 | miRNA 21 UCCCUAACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a-3p | AcCAT6 | 3.0 | miRNA 21 CCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a-3p | AcCAT5 | 3.0 | miRNA 21 CCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166b | AcCAT6 | 3.0 | miRNA 21 AUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166b | AcCAT5 | 3.0 | miRNA 21 AUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166g-3p | AcCAT5 | 3.0 | miRNA 21 CUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166g-3p | AcCAT6 | 3.0 | miRNA 21 CUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166m | AcCAT5 | 3.0 | miRNA 21 UCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166m | AcCAT6 | 3.0 | miRNA 21 UCCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166n | AcCAT5 | 3.0 | miRNA 21 UUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1338 AUGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166n | AcCAT6 | 3.0 | miRNA 21 UUCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : : :. : : Target 1449 AUGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a | AcCAT6 | 3.0 | miRNA 19 CCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1451 GGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
| ach-miR166a | AcCAT5 | 3.0 | miRNA 19 CCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1340 GGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166h-3p | AcCAT5 | 3.0 | miRNA 20 CCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1339 UGGAUUGAAGCCUUGUCUGA 1358 | 剪切 Cleavage |
| ach-miR166h-3p | AcCAT6 | 3.0 | miRNA 20 CCCUUACUUCGGACCAGGCU 1 : : : : : : : : : : : : : :. : : Target 1450 UGGAUUGAAGCCUUGUCUGA 1469 | 剪切 Cleavage |
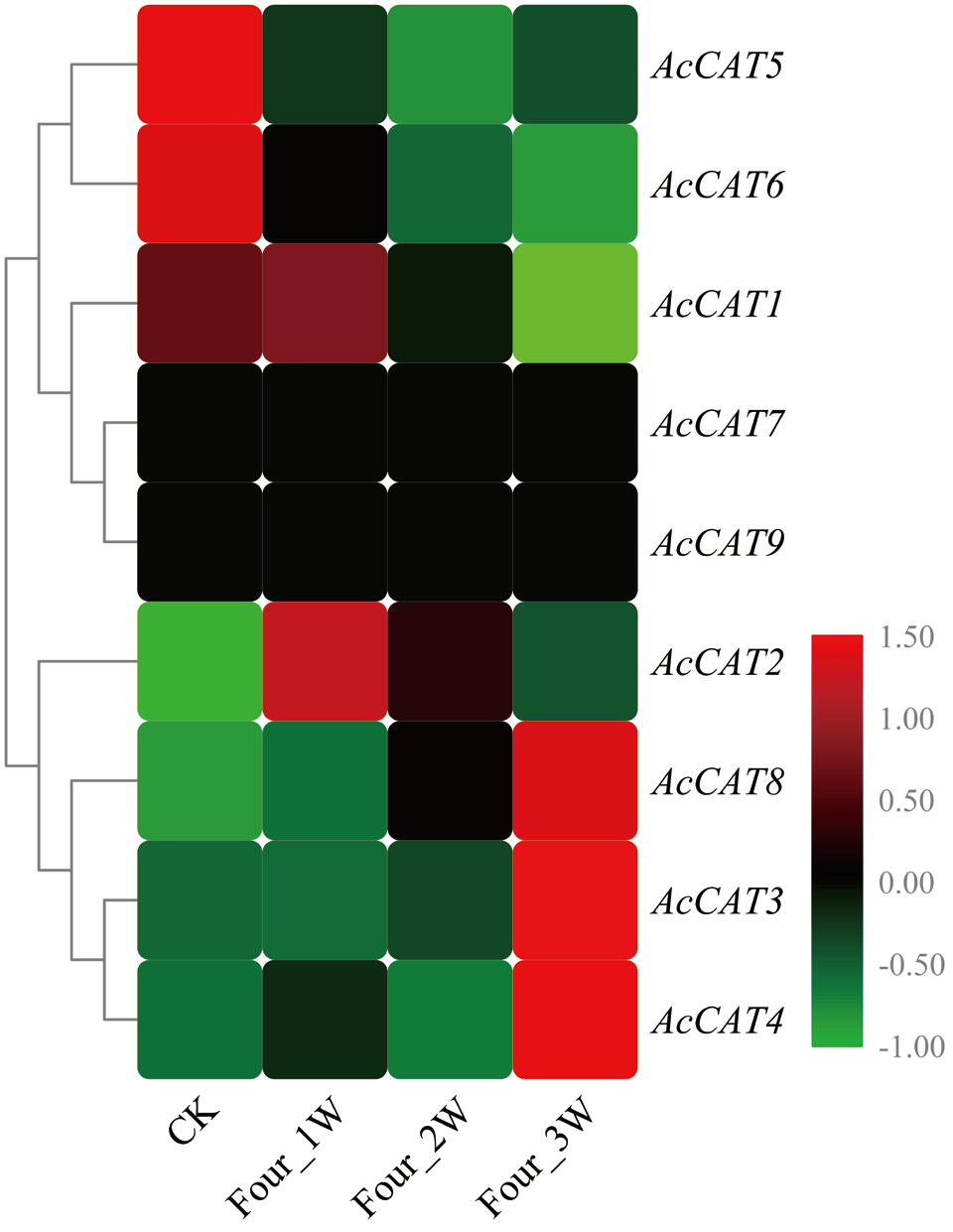
图8 猕猴桃AcCAT基因在果实贮藏过程中的表达谱 CK表示贮藏0 d,Four_1W为4℃条件下贮藏1周,Four_2W为4℃条件下贮藏2周,Four_3W为4℃条件下贮藏3周。下同
Fig. 8 Expression profile of AcCAT genes during the storage of kiwifruit CK indicates control check, Four_1W, Four_2W, Four_3W indicates kiwifruit stored in 4℃ for 1, 2 and 3 week, respectively. The same below
| miRNA | AcCAT5 | AcCAT6 | ||||
|---|---|---|---|---|---|---|
| 皮尔逊相关性Pearson correlation | 显著性Significance | 皮尔逊相关性Pearson correlation | 显著性Significance | |||
| ach-miR166k-3p | -0.291 | 0.709 | -0.495 | 0.505 | ||
| ach-miR166a-3p | -0.479 | 0.521 | -0.651 | 0.349 | ||
| ach-miR166b | -0.990* | 0.010 | -0.976* | 0.024 | ||
| ach-miR166g-3p | -0.996** | 0.004 | -0.975* | 0.025 | ||
| ach-miR166m | -0.069 | 0.931 | -0.259 | 0.741 | ||
| ach-miR166n | -0.991** | 0.009 | -0.978* | 0.022 | ||
| ach-miR166a | -0.989* | 0.011 | -0.976* | 0.024 | ||
| ach-miR166h-3p | -0.482 | 0.518 | -0.653 | 0.347 | ||
表3 基于表达谱的猕猴桃AcCAT基因与miR166家族成员的相关性分析
Table 3 Correlation analysis of AcCAT genes and miR166 family members in A. chinensis based on expression profile
| miRNA | AcCAT5 | AcCAT6 | ||||
|---|---|---|---|---|---|---|
| 皮尔逊相关性Pearson correlation | 显著性Significance | 皮尔逊相关性Pearson correlation | 显著性Significance | |||
| ach-miR166k-3p | -0.291 | 0.709 | -0.495 | 0.505 | ||
| ach-miR166a-3p | -0.479 | 0.521 | -0.651 | 0.349 | ||
| ach-miR166b | -0.990* | 0.010 | -0.976* | 0.024 | ||
| ach-miR166g-3p | -0.996** | 0.004 | -0.975* | 0.025 | ||
| ach-miR166m | -0.069 | 0.931 | -0.259 | 0.741 | ||
| ach-miR166n | -0.991** | 0.009 | -0.978* | 0.022 | ||
| ach-miR166a | -0.989* | 0.011 | -0.976* | 0.024 | ||
| ach-miR166h-3p | -0.482 | 0.518 | -0.653 | 0.347 | ||
| [1] |
Nie Q, Gao GL, Fan QJ, et al. Isolation and characterization of a catalase gene “HuCAT3” from pitaya(Hylocereus undatus)and its expression under abiotic stress[J]. Gene, 2015, 563(1): 63-71.
doi: 10.1016/j.gene.2015.03.007 URL |
| [2] |
Sofo A, Scopa A, Nuzzaci M, et al. Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses[J]. Int J Mol Sci, 2015, 16(6): 13561-13578.
doi: 10.3390/ijms160613561 pmid: 26075872 |
| [3] |
Chen HJ, Wu SD, Huang GJ, et al. Expression of a cloned sweet potato catalase SPCAT1 alleviates ethephon-mediated leaf senescence and H2O2 elevation[J]. J Plant Physiol, 2012, 169(1): 86-97.
doi: 10.1016/j.jplph.2011.08.002 URL |
| [4] |
Du YY, Wang PC, Chen J, et al. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana[J]. J Integr Plant Biol, 2008, 50(10): 1318-1326.
doi: 10.1111/jipb.2008.50.issue-10 URL |
| [5] |
Joo J, Lee YH, Song SI. Rice CatA, CatB, and CatC are involved in environmental stress response, root growth, and photorespiration, respectively[J]. J Plant Biol, 2014, 57(6): 375-382.
doi: 10.1007/s12374-014-0383-8 URL |
| [6] |
Wang W, Cheng YY, Chen DD, et al. The catalase gene family in cotton: genome-wide characterization and bioinformatics analysis[J]. Cells, 2019, 8(2): 86.
doi: 10.3390/cells8020086 URL |
| [7] |
Zhang Y, Zheng LJ, Yun L, et al. Catalase(CAT)gene family in wheat(Triticum aestivum L.): evolution, expression pattern and function analysis[J]. Int J Mol Sci, 2022, 23(1): 542.
doi: 10.3390/ijms23010542 URL |
| [8] |
Raza A, Su W, Gao A, et al. Catalase(CAT)gene family in rapeseed(Brassica napus L.): genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions[J]. Int J Mol Sci, 2021, 22(8): 4281.
doi: 10.3390/ijms22084281 URL |
| [9] |
Purev M, Kim YJ, Kim MK, et al. Isolation of a novel catalase(Cat1)gene from Panax ginseng and analysis of the response of this gene to various stresses[J]. Plant Physiol Biochem, 2010, 48(6): 451-460.
doi: 10.1016/j.plaphy.2010.02.005 URL |
| [10] |
Li YF, Jiang WJ, Liu CH, et al. Comparison of fruit morphology and nutrition metabolism in different cultivars of kiwifruit across developmental stages[J]. PeerJ, 2021, 9: e11538.
doi: 10.7717/peerj.11538 URL |
| [11] |
Liu YF, Qi YW, Chen X, et al. Phenolic compounds and antioxidant activity in red- and in green-fleshed kiwifruits[J]. Food Res Int, 2019, 116: 291-301.
doi: S0963-9969(18)30653-7 pmid: 30716948 |
| [12] |
Pan LY, Zhao XY, Chen M, et al. Effect of exogenous methyl jasmonate treatment on disease resistance of postharvest kiwifruit[J]. Food Chem, 2020, 305: 125483.
doi: 10.1016/j.foodchem.2019.125483 URL |
| [13] |
Xu FX, Liu SY, Liu YF, et al. Effectiveness of lysozyme coatings and 1-MCP treatments on storage and preservation of kiwifruit[J]. Food Chem, 2019, 288: 201-207.
doi: S0308-8146(19)30490-X pmid: 30902282 |
| [14] |
千春录, 殷建东, 王利斌, 等. 1-甲基环丙烯和自发气调对猕猴桃品质及活性氧代谢的影响[J]. 食品科学, 2018, 39(11): 233-240.
doi: 10.7506/spkx1002-6630-201811037 |
| Qian CL, Yin JD, Wang LB, et al. Effects of 1-methylcyclopropene treatment and self-developed modified atmosphere on quality and reactive oxygen species metabolism of kiwifruits during storage[J]. Food Sci, 2018, 39(11): 233-240. | |
| [15] |
梁春强, 吕茳, 靳蜜静, 等. 草酸处理对采后猕猴桃冷害、抗氧化能力及能荷的影响[J]. 园艺学报, 2017, 44(2): 279-287.
doi: 10.16420/j.issn.0513-353x.2016-0451 |
| Liang CQ, Lv J, Jin MJ, et al. Effects of oxalic acid treatment on chilling injury, antioxidant capacity and energy status in harvested kiwifruits under low temperature stress[J]. Acta Hortic Sin, 2017, 44(2): 279-287. | |
| [16] |
张承, 李明, 龙友华, 等. 采前喷施壳聚糖复合膜对猕猴桃软腐病的防控及其保鲜作用[J]. 食品科学, 2016, 37(22): 274-281.
doi: 10.7506/spkx1002-6630-201622042 |
|
Zhang C, Li M, Long YH, et al. Control of soft rot in kiwifruit by pre-harvest application of chitosan composite coating and its effect on preserving and improving kiwifruit quality[J]. Food Sci, 2016, 37(22): 274-281.
doi: 10.1111/jfds.1972.37.issue-2 URL |
|
| [17] | 沈勇根, 汪伟, 张伟, 等. 硫化氢提高低温贮藏下猕猴桃的抗氧化能力及果实品质(英文)[J]. 农业工程学报, 2015, 31(S1): 367-372. |
| Shen YG, Wang W, Zhang W, et al. Hydrogen sulfide facilitating enhancement of antioxidant ability and maintainance of fruit quality of kiwifruits during low-temperature storage[J]. Trans Chin Soc Agric Eng, 2015, 31(S1): 367-372. | |
| [18] |
Liang D, Shen YQ, Ni ZY, et al. Exogenous melatonin application delays senescence of kiwifruit leaves by regulating the antioxidant capacity and biosynthesis of flavonoids[J]. Front Plant Sci, 2018, 9: 426.
doi: 10.3389/fpls.2018.00426 pmid: 29675031 |
| [19] |
Liang D, Gao F, Ni ZY, et al. Melatonin improves heat tolerance in kiwifruit seedlings through promoting antioxidant enzymatic activity and glutathione S-transferase transcription[J]. Molecules, 2018, 23(3): 584.
doi: 10.3390/molecules23030584 URL |
| [20] |
Xia H, Ni ZY, Hu RP, et al. Melatonin alleviates drought stress by a non-enzymatic and enzymatic antioxidative system in kiwifruit seedlings[J]. Int J Mol Sci, 2020, 21(3): 852.
doi: 10.3390/ijms21030852 URL |
| [21] | Blokhina O, Virolainen E, Fagerstedt KV. Antioxidants, oxidative damage and oxygen deprivation stress: a review[J]. Ann Bot, 2003, 91 Spec No(2): 179-194. |
| [22] | Wei CL, Yang H, Wang SB, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality[J]. Proc Natl Acad Sci USA, 2018, 115(18): E4151-E4158. |
| [23] |
Li XH, Liu GY, Geng YH, et al. A genome-wide analysis of the small auxin-up RNA(SAUR)gene family in cotton[J]. BMC Genomics, 2017, 18(1): 815.
doi: 10.1186/s12864-017-4224-2 URL |
| [24] |
Song ZP, Pan FL, Lou XP, et al. Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum[J]. Mol Biol Rep, 2019, 46(2): 1941-1954.
doi: 10.1007/s11033-019-04644-7 |
| [25] |
Xu XP, Chen XH, Shen X, et al. Genome-wide identification and characterization of DEAD-box helicase family associated with early somatic embryogenesis in Dimocarpus longan Lour[J]. J Plant Physiol, 2021, 258/259: 153364.
doi: 10.1016/j.jplph.2021.153364 URL |
| [26] |
Xu K, Zhao Y, Zhao SH, et al. Genome-wide identification and low temperature responsive pattern of actin depolymerizing factor(ADF)gene family in wheat(Triticum aestivum L.)[J]. Front Plant Sci, 2021, 12: 618984.
doi: 10.3389/fpls.2021.618984 URL |
| [27] |
Zhao W, Liu YH, Li L, et al. Genome-wide identification and characterization of bHLH transcription factors related to anthocyanin biosynthesis in red walnut(Juglans regia L.)[J]. Front Genet, 2021, 12: 632509.
doi: 10.3389/fgene.2021.632509 URL |
| [28] |
Zhang ZS, Chen J, Liang CL, et al. Genome-wide identification and characterization of the bHLH transcription factor family in pepper(Capsicum annuum L.)[J]. Front Genet, 2020, 11: 570156.
doi: 10.3389/fgene.2020.570156 URL |
| [29] |
Cheng CZ, Liu F, Sun XL, et al. Genome-wide identification of FAD gene family and their contributions to the temperature stresses and mutualistic and parasitic fungi colonization responses in banana[J]. Int J Biol Macromol, 2022, 204: 661-676.
doi: 10.1016/j.ijbiomac.2022.02.024 URL |
| [30] |
李濯雪, 陈信波. 植物诱导型启动子及相关顺式作用元件研究进展[J]. 生物技术通报, 2015, 31(10): 8-15.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.10.006 |
| Li ZX, Chen XB. Research advances on plant inducible promoters and related cis-acting elements[J]. Biotechnol Bull, 2015, 31(10): 8-15. | |
| [31] |
Sun ZC, Shu LL, Zhang W, et al. Cca-miR398 increases copper sulfate stress sensitivity via the regulation of CSD mRNA transcription levels in transgenic Arabidopsis thaliana[J]. PeerJ, 2020, 8: e9105.
doi: 10.7717/peerj.9105 URL |
| [32] |
Liu F, Huang N, Wang L, et al. A novel L-ascorbate peroxidase 6 gene, ScAPX6, plays an important role in the regulation of response to biotic and abiotic stresses in sugarcane[J]. Front Plant Sci, 2018, 8: 2262.
doi: 10.3389/fpls.2017.02262 URL |
| [33] |
Bela K, Horváth E, Gallé Á, et al. Plant glutathione peroxidases: emerging role of the antioxidant enzymes in plant development and stress responses[J]. J Plant Physiol, 2015, 176: 192-201.
doi: 10.1016/j.jplph.2014.12.014 URL |
| [34] |
Begara-Morales JC, Sánchez-Calvo B, Chaki M, et al. Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S-nitrosylation[J]. J Exp Bot, 2015, 66(19): 5983-5996.
doi: 10.1093/jxb/erv306 pmid: 26116026 |
| [35] |
Kitazumi A, Kawahara Y, Onda TS, et al. Implications of miR166 and miR159 induction to the basal response mechanisms of an Andigena potato(Solanum tuberosum subsp. andigena)to salinity stress, predicted from network models in Arabidopsis[J]. Genome, 2015, 58(1): 13-24.
doi: 10.1139/gen-2015-0011 pmid: 25955479 |
| [36] |
Ding YF, Gong SH, Wang Y, et al. microRNA166 modulates cadmium tolerance and accumulation in rice[J]. Plant Physiol, 2018, 177(4): 1691-1703.
doi: 10.1104/pp.18.00485 pmid: 29925586 |
| [37] |
Li N, Yang TX, Guo ZY, et al. Maize microRNA166 inactivation confers plant development and abiotic stress resistance[J]. Int J Mol Sci, 2020, 21(24): 9506.
doi: 10.3390/ijms21249506 URL |
| [38] |
Ravichandran S, Ragupathy R, Edwards T, et al. microRNA-guided regulation of heat stress response in wheat[J]. BMC Genomics, 2019, 20(1): 488.
doi: 10.1186/s12864-019-5799-6 pmid: 31195958 |
| [39] |
Li XY, Xie X, Li J, et al. Conservation and diversification of the miR166 family in soybean and potential roles of newly identified miR166s[J]. BMC Plant Biol, 2017, 17(1): 32.
doi: 10.1186/s12870-017-0983-9 pmid: 28143404 |
| [40] |
Hamza NB, Sharma N, Tripathi A, et al. microRNA expression profiles in response to drought stress in Sorghum bicolor[J]. Gene Expr Patterns, 2016, 20(2): 88-98.
doi: 10.1016/j.gep.2016.01.001 URL |
| [1] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [2] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [3] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [4] | 刘奎, 李兴芬, 杨沛欣, 仲昭晨, 曹一博, 张凌云. 青杄转录共激活因子PwMBF1c的功能研究与验证[J]. 生物技术通报, 2023, 39(5): 205-216. |
| [5] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [6] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [7] | 姚晓文, 梁晓, 陈青, 伍春玲, 刘迎, 刘小强, 税军, 乔阳, 毛奕茗, 陈银华, 张银东. 二斑叶螨抗性木薯木质素合成途径基因表达特性研究[J]. 生物技术通报, 2023, 39(2): 161-171. |
| [8] | 李彦霞, 王晋鹏, 冯芬, 包斌武, 董益闻, 王兴平, 罗仍卓么. 大肠杆菌型奶牛乳房炎对产奶性状相关基因表达的影响[J]. 生物技术通报, 2023, 39(2): 274-282. |
| [9] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| [10] | 毛可欣, 王海荣, 安淼, 刘腾飞, 王世金, 李健, 李国田. 中华猕猴桃GRAS基因家族鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 297-307. |
| [11] | 吴柏增, 何琪, 姚方杰, 赵梦然. 糙皮侧耳乳酸脱氢酶鉴定及其菌丝高温胁迫下表达特征分析[J]. 生物技术通报, 2023, 39(11): 350-359. |
| [12] | 姜南, 石杨, 赵志慧, 李斌, 赵熠辉, 杨俊彪, 闫家铭, 靳雨璠, 陈稷, 黄进. 镉胁迫下水稻OsPT1的表达及功能分析[J]. 生物技术通报, 2023, 39(1): 166-174. |
| [13] | 段敏杰, 李怡斐, 杨小苗, 王春萍, 黄启中, 黄任中, 张世才. 辣椒锌指蛋白DnaJ-Like基因家族鉴定及对高温胁迫的表达响应[J]. 生物技术通报, 2023, 39(1): 187-198. |
| [14] | 袁星, 郭彩华, 刘金明, 亢超, 全绍文, 牛建新. 核桃CONSTANS-Like基因家族全基因组鉴定及表达分析[J]. 生物技术通报, 2022, 38(9): 167-179. |
| [15] | 郭宾会, 宋丽. 大豆孢囊线虫侵染对乙烯合成及信号传导基因表达调控的研究[J]. 生物技术通报, 2022, 38(8): 150-158. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||