








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 246-258.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1045
王艺清1,2( ), 王涛1,2, 韦朝领3, 戴浩民1,2, 曹士先4, 孙威江1,2(
), 王涛1,2, 韦朝领3, 戴浩民1,2, 曹士先4, 孙威江1,2( ), 曾雯1,2(
), 曾雯1,2( )
)
收稿日期:2022-08-23
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
孙威江,男,博士,教授,研究方向:茶树种质资源创新与品质化学;E-mail: swj8103@126.com;作者简介:王艺清,女,硕士研究生,研究方向:茶树分子生物学;E-mail: 784726205@qq.com
基金资助:
WANG Yi-qing1,2( ), WANG Tao1,2, WEI Chao-ling3, DAI Hao-min1,2, CAO Shi-xian4, SUN Wei-jiang1,2(
), WANG Tao1,2, WEI Chao-ling3, DAI Hao-min1,2, CAO Shi-xian4, SUN Wei-jiang1,2( ), ZENG Wen1,2(
), ZENG Wen1,2( )
)
Received:2022-08-23
Published:2023-04-26
Online:2023-05-16
摘要:
S-腺苷甲硫氨酸合成酶(S-adenosylmethionine synthase, SAMS)是催化甲硫氨酸和ATP合成S-腺苷甲硫氨酸(SAM)的唯一酶,研究表明SAMS参与木质素的生物合成。本研究对‘黄棪’茶树(Camellia sinensis)中鉴定的SAMS基因家族,进行表达模式及蛋白互作网络分析,挖掘可能参与木质素合成的 CsSAMS候选基因。以‘黄棪’茶树基因组为参考基因组,通过生物信息学鉴定CsSAMS基因家族成员,并分析其蛋白理化性质、系统进化树、染色体定位、基因结构、蛋白结构、表达模式,通过酵母双杂交技术(Y2H)研究其蛋白互作网络,通过紫外分光光度计方法对‘黄棪’‘铁观音’‘金观音’‘福鼎大毫茶’一芽二叶部位的木质素含量进行测定。生物信息学分析结果表明,‘黄棪’茶树中共鉴定到4个CsSAMS家族成员,其编码氨基酸个数为345-519,等电点为6.12-6.47。亚细胞定位预测结果表明,CsSAMS1定位于叶绿体,CsSAMS2、CsSAMS3定位于细胞质,CsSAMS4定位于细胞骨架。通过对不同茶树品种的CsSAMS表达量和木质素含量检测发现,CsSAMS2、CsSAMS3、CsSAMS4可能潜在调控木质素的含量,另外酵母双杂交结果表明,CsSAMS4可以与自身形成同源二聚体。本研究鉴定并分析了4个CsSAMS成员的理化性质并预测其功能,明确了不同组织部位和氮、氟处理下CsSAMS基因的表达模式,以及CsSAMS对木质素合成过程的潜在参与作用。
王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258.
WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis)[J]. Biotechnology Bulletin, 2023, 39(4): 246-258.
| 基因名称Gene name | 引物序列Primer sequence(5'-3') | 备注Note |
|---|---|---|
| SAMS1 | F CATGCCCAAGTGTAACAATAG R AGAGACGGAGAGAGAGAAGA | 荧光定量 Quantitative real-time PCR |
| SAMS2 | F CCACTGATGAAACACCTGAAC R GTATGAACTCTAATGGGGACC | |
| SAMS3 | F CACGGGGTTCTTTCTCTCTGTCT R GGTGTCCCTCATTCACTGATTCA | |
| SAMS4 | F AGCCCGTCATCCCTTCCCAATA R CTCCACCGTGAGCACCCCAACC | |
| BD- SAMS2 | F CATGGAGGCCGAATTCATGGATAGTTTCCTC R GCAGGTCGACGGATCCTCAAGCTTTGGGCT | 酵母双杂交 Yeast two-hybrid |
| BD- SAMS4 | F CATGGAGGCCGAATTCATGGAAACCTTCCTCT R GCAGGTCGACGGATCCTCAAGCTTTGGGCT | |
| AD- SAMS2 | F CATCGATACGGGATCATGGATAGTTTCCTCTT R ACGATTCATCTGCAGCTCAAGCTTTGGGCTTAA | |
| AD- SAMS4 | F CATCGATACGGGATCATGGAAACCTTCCTCT R ACGATTCATCTGCAGCTCAAGCTTTGGGCTTG |
表1 引物序列
Table 1 Primer sequences
| 基因名称Gene name | 引物序列Primer sequence(5'-3') | 备注Note |
|---|---|---|
| SAMS1 | F CATGCCCAAGTGTAACAATAG R AGAGACGGAGAGAGAGAAGA | 荧光定量 Quantitative real-time PCR |
| SAMS2 | F CCACTGATGAAACACCTGAAC R GTATGAACTCTAATGGGGACC | |
| SAMS3 | F CACGGGGTTCTTTCTCTCTGTCT R GGTGTCCCTCATTCACTGATTCA | |
| SAMS4 | F AGCCCGTCATCCCTTCCCAATA R CTCCACCGTGAGCACCCCAACC | |
| BD- SAMS2 | F CATGGAGGCCGAATTCATGGATAGTTTCCTC R GCAGGTCGACGGATCCTCAAGCTTTGGGCT | 酵母双杂交 Yeast two-hybrid |
| BD- SAMS4 | F CATGGAGGCCGAATTCATGGAAACCTTCCTCT R GCAGGTCGACGGATCCTCAAGCTTTGGGCT | |
| AD- SAMS2 | F CATCGATACGGGATCATGGATAGTTTCCTCTT R ACGATTCATCTGCAGCTCAAGCTTTGGGCTTAA | |
| AD- SAMS4 | F CATCGATACGGGATCATGGAAACCTTCCTCT R ACGATTCATCTGCAGCTCAAGCTTTGGGCTTG |
| 基因 Gene | 序列号 Gene ID | 蛋白长度 Amino acid/aa | 分子量 Molecular weight/kD | 等电点 pI | 不稳定系数 Instability constant | 亲水性 GRAVY | 亚细胞定位 Subcellular localization | 跨膜结构 Transmembrane domain | 信号肽 Signal peptide |
|---|---|---|---|---|---|---|---|---|---|
| CsSAMS1 | HD.05G0021870 | 519 | 56.98 | 6.47 | 28.69 | -0.245 | 叶绿体Chloroplast | 无 No | 无 No |
| CsSAMS2 | HD.06G0008900 | 390 | 42.75 | 6.24 | 25.34 | -0.331 | 细胞质Cytoplasm | 无 No | 无 No |
| CsSAMS3 | HD.08G0000160 | 345 | 37.79 | 6.42 | 18.7 | -0.281 | 细胞质Cytoplasm | 无 No | 无 No |
| CsSAMS4 | HD.11G0014340 | 390 | 42.68 | 6.12 | 27.59 | -0.294 | 细胞骨架Cytoskeleton | 无 No | 无 No |
表2 CsSAMS蛋白的基本理化性质
Table 2 Basic physicochemical properties of CsSAMS proteins
| 基因 Gene | 序列号 Gene ID | 蛋白长度 Amino acid/aa | 分子量 Molecular weight/kD | 等电点 pI | 不稳定系数 Instability constant | 亲水性 GRAVY | 亚细胞定位 Subcellular localization | 跨膜结构 Transmembrane domain | 信号肽 Signal peptide |
|---|---|---|---|---|---|---|---|---|---|
| CsSAMS1 | HD.05G0021870 | 519 | 56.98 | 6.47 | 28.69 | -0.245 | 叶绿体Chloroplast | 无 No | 无 No |
| CsSAMS2 | HD.06G0008900 | 390 | 42.75 | 6.24 | 25.34 | -0.331 | 细胞质Cytoplasm | 无 No | 无 No |
| CsSAMS3 | HD.08G0000160 | 345 | 37.79 | 6.42 | 18.7 | -0.281 | 细胞质Cytoplasm | 无 No | 无 No |
| CsSAMS4 | HD.11G0014340 | 390 | 42.68 | 6.12 | 27.59 | -0.294 | 细胞骨架Cytoskeleton | 无 No | 无 No |

图3 CsSAMS系统进化树分析
Fig. 3 Phylogene tree of CsSAMS CsSAM1-4:茶树 Camellia sinensis;AtMAT1(At1g02500)、AtMAT2(At4g01850)、AtMAT3(At2g36880)AtMAT4(At3g17390):拟南芥 Arabidopsis thaliana;SlSAMS1(NP.001234425)、SlSAMS2(NP.001296305)、SlSAMS3(NP.001234004)、SlSAMS4(XP.010312254):番茄 Solanum lycopersicum;ZmSAMS1(NP.001130734)、ZmSAMS2(NP.001146249)、ZmSAMS3(NP.001148708)、ZmSAMS4(NP.001132867):玉米 Zea mays;MtSAMS1(XP.003626035)、MtSAMS2(XP.003609861)、MtSAMS3a(XP.003625682)、MtSAMS3b(XP.013468134)、MtSAMS4(XP.013463713):蒺藜苜蓿 Medicago truncatula;TuSAMS(XP.020178978)、TuSAMS1(EMS55466)、TuSAMS4(EMS52834):乌拉尔图小麦 Triticum urartu;OsSAMS1(NP.001389410)、OsSAMS2(NP.001393223)、OsSAMS3(XP.015614349):水稻 Oryza sativa;DcSAMS(AAA33274):康乃馨 Dianthus caryophyllus;LcSAMS(AAP13994):荔枝 Litchi chinensis;PcSAMS(AAG17036):扭叶松 Pinus contorta;RpSAMS(AIT39705):刺槐 Robinia pseudoacacia;LrSAMS(AFC88125):石蒜 Lycoris radiata;BjSAMS(AAK71234):芥菜 Brassica juncea;CcSAMS(KYP71740):木豆 Cajanus cajan
| 基因对名称 Gene pair name | 非同义突变频率Nonsynonymous mutation Ka | 同义突变频率Synonymous mutation Ks | Ka/Ks |
|---|---|---|---|
| CsSAMS1/CsSAMS3 | 0.033 115 095 | 1.036 155 399 | 0.031 959 584 |
| CsSAMS2/CsSAMS4 | 0.022 137 640 | 0.526 911 233 | 0.042 013 983 |
表3 CsSAMS同源基因的Ka/Ks分析
Table 3 Ka/Ks analysis of CsSAMS homologous genes
| 基因对名称 Gene pair name | 非同义突变频率Nonsynonymous mutation Ka | 同义突变频率Synonymous mutation Ks | Ka/Ks |
|---|---|---|---|
| CsSAMS1/CsSAMS3 | 0.033 115 095 | 1.036 155 399 | 0.031 959 584 |
| CsSAMS2/CsSAMS4 | 0.022 137 640 | 0.526 911 233 | 0.042 013 983 |
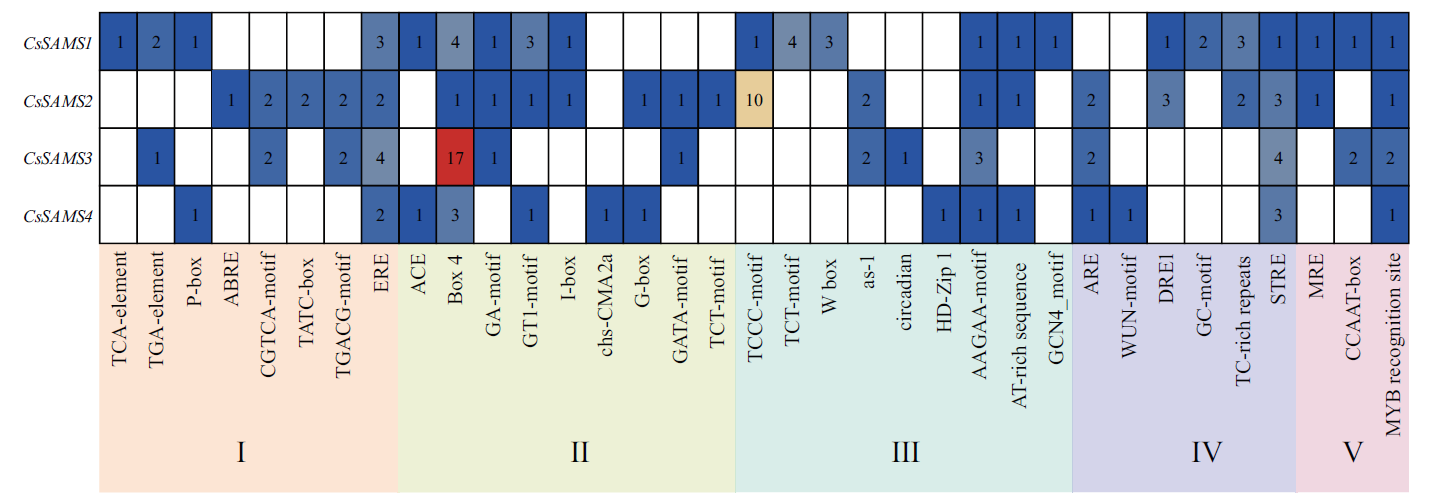
图6 CsSAMS启动子顺式作用元件预测 I:激素响应;Ⅱ:非生物胁迫响应;Ⅲ:光响应;Ⅳ:转录因子识别、结合位点;Ⅴ:组织特异性元件;Ⅵ:生长发育相关元件;Ⅶ:核心元件
Fig. 6 Analysis of cis-elements in the upstream region of CsSAMS family genes in tea plants I: Phytohormone response. Ⅱ: Abiotic stress response. Ⅲ: Light response. Ⅳ: TF recongnition and binding site. Ⅴ: Tissue specificity. Ⅵ: Plant growth. Ⅶ: Core
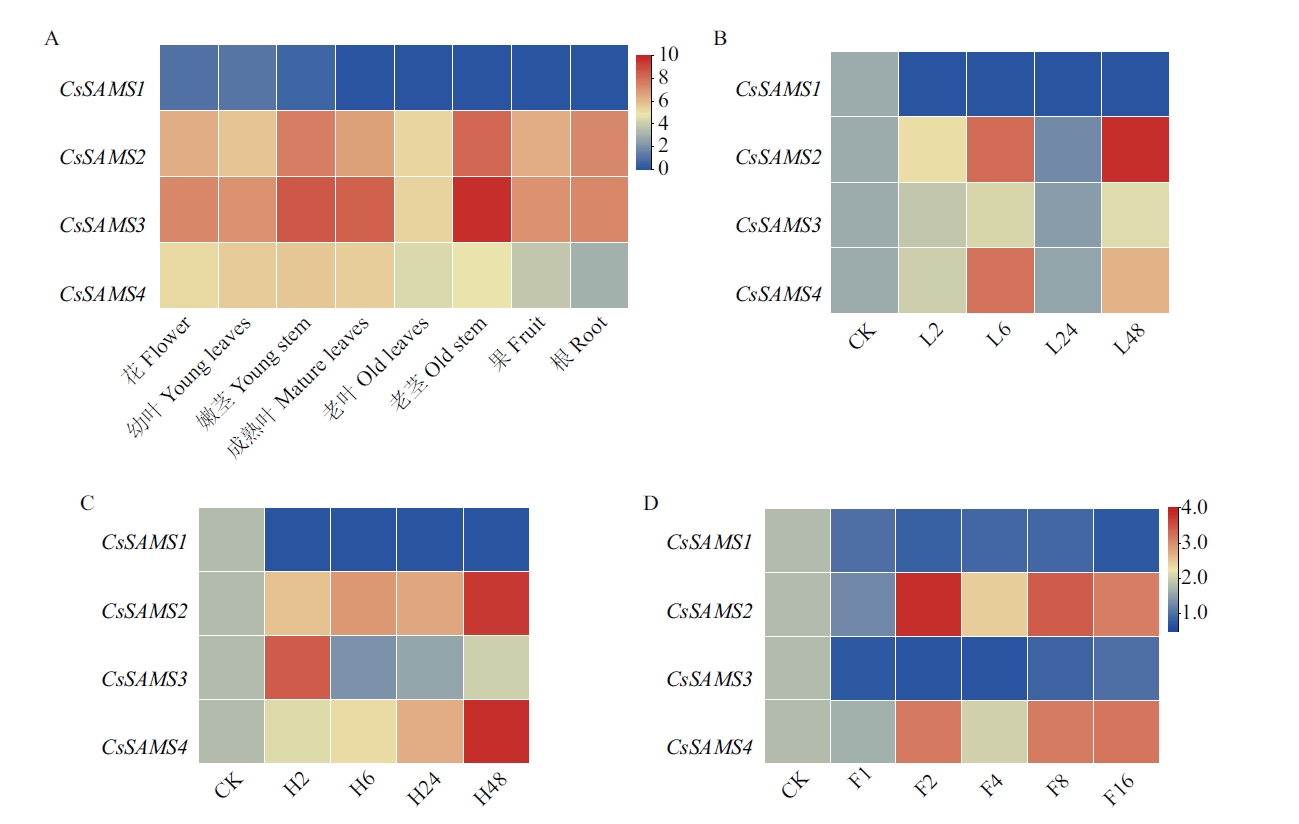
图8 CsSAMS基因表达模式 A:CsSAMS 在不同组织部位中的表达模式;B:CsSAMS 在低氮处理下的表达模式;C:CsSAMS 在高氮处理下的表达模式;D:CsSAMS 在氟处理下的表达模式
Fig. 8 Expression patterns of CsSAMS A: Expression pattern of CsSAMS in different tissue. B: Expression pattern of CsSAMS under low nitrogen treatment. C: Expression pattern of CsSAMS under high nitrogen treatment. D: Expression pattern of CsSAMS under fluorine treatment

图11 不同茶树品种木质素含量和CsSAMS表达量相关性分析 *表示在0.05水平上显著相关,**表示在0.01水平上显著相关
Fig. 11 Correlation analysis between the lignin content and CsSAMS genes expression in different tea plant varieties * indicates significant correlation at the 0.05 level, and ** indicates significant correlation at the 0.01 level
| [1] |
Vanholme R, de Meester B, Ralph J, et al. Lignin biosynthesis and its integration into metabolism[J]. Curr Opin Biotechnol, 2019, 56: 230-239.
doi: 10.1016/j.copbio.2019.02.018 URL |
| [2] | 王永鑫. 茶树木质素代谢分子机制研究[D]. 南京: 南京农业大学, 2019. |
| Wang YX. Study of molecular mechanism of lignin metabolism in tea plant(Camellia sinensis)[D]. Nanjing: Nanjing Agricultural University, 2019. | |
| [3] |
Wang YX, Teng RM, Wang WL, et al. Identification of genes reveal-ed differential expression profiles and lignin accumulation during leaf and stem development in tea plant(Camellia sinensis(L.) O. Kuntze)[J]. Protoplasma, 2019, 256(2): 359-370.
doi: 10.1007/s00709-018-1299-9 |
| [4] | 李寿田, 周健民, 朱世东, 等. 萝卜贮藏期间木质素、纤维素和可溶性糖含量变化及其与糠心的关系[J]. 安徽农业大学学报, 2001, 28(3): 255-258. |
| Li ST, Zhou JM, Zhu SD, et al. Content changes of lignin, cellulose and soluble sugar and their correlations with hollowness during storage in radishes[J]. J Anhui Agric Univ, 2001, 28(3): 255-258. | |
| [5] | Bedon F, Legay S. Lignin synthesis, transcriptional regulation and potential for useful modification in plants[J]. CABI Rev, 2011, 2011: 1-28. |
| [6] | 杨建坤. 木质素与茶树新梢嫩度关系的研究[D]. 杨凌: 西北农林科技大学, 2019. |
| Yang JK. Study on relation between lignin and shoot tendness of Camellia sinensis[D]. Yangling: Northwest A & F University, 2019. | |
| [7] |
Dixon RA, Barros J. Lignin biosynthesis: old roads revisited and new roads explored[J]. Open Biol, 2019, 9(12): 190215.
doi: 10.1098/rsob.190215 URL |
| [8] |
Monné M, Marobbio CMT, Agrimi G, et al. Mitochondrial transport and metabolism of the major methyl donor and versatile cofactor S-adenosylmethionine, and related diseases: a review[J]. IUBMB Life, 2022, 74(7): 573-591.
doi: 10.1002/iub.2658 pmid: 35730628 |
| [9] |
Hanson AD, Roje S. One-carbon metabolism in higher plants[J]. Annu Rev Plant Physiol Plant Mol Biol, 2001, 52: 119-137.
doi: 10.1146/arplant.2001.52.issue-1 URL |
| [10] | Li YY, Ogita S, Keya CA, et al. Expression of caffeine biosynthesis genes in tea(Camellia sinensis)[J]. Z Naturforsch C J Biosci, 2008, 63(3/4): 267-270. |
| [11] |
吕焕青, 王志敏, 汤青林, 等. 多胺生物合成途径中两个关键酶基因研究进展[J]. 生物技术通报, 2015, 31(2): 61-64.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.02.008 |
|
Lü HQ, Wang ZM, Tang QL, et al. Polyamine biosynthesis enzyme research progress in two key genes[J]. Biotechnol Bull, 2015, 31(2): 61-64.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.02.008 |
|
| [12] |
Roje S. S-Adenosyl-L-methionine: beyond the universal methyl group donor[J]. Phytochemistry, 2006, 67(15): 1686-1698.
doi: 10.1016/j.phytochem.2006.04.019 pmid: 16766004 |
| [13] |
Sekula B, Ruszkowski M, Dauter Z. S-adenosylmethionine synthases in plants: structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula[J]. Int J Biol Macromol, 2020, 151: 554-565.
doi: 10.1016/j.ijbiomac.2020.02.100 URL |
| [14] |
Shen B, Li CJ, Tarczynski MC. High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-L-methionine synthetase 3 gene[J]. Plant J, 2002, 29(3): 371-380.
doi: 10.1046/j.1365-313x.2002.01221.x pmid: 11844113 |
| [15] |
Meng JJ, Wang LS, Wang JY, et al. METHIONINE ADENOSYLTRANSFERASE4 mediates DNA and histone methylation[J]. Plant Physiol, 2018, 177(2): 652-670.
doi: 10.1104/pp.18.00183 pmid: 29572390 |
| [16] |
Yang SX, Wu TT, Ding CH, et al. SAHH and SAMS form a methyl donor complex with CCoAOMT7 for methylation of phenolic compounds[J]. Biochem Biophys Res Commun, 2019, 520(1): 122-127.
doi: 10.1016/j.bbrc.2019.09.101 URL |
| [17] |
Li Y, Xiong WD, He F, et al. Down-regulation of PvSAMS impairs S-adenosyl-L-methionine and lignin biosynthesis, and improves cell wall digestibility in switchgrass[J]. J Exp Bot, 2022, 73(12): 4157-4169.
doi: 10.1093/jxb/erac147 URL |
| [18] |
Yang X, Yu Z, Zhang BB, et al. Effect of fluoride on the biosynthesis of catechins in tea[Camellia sinensis(L.) O. Kuntze]leaves[J]. Sci Hortic, 2015, 184: 78-84.
doi: 10.1016/j.scienta.2014.12.031 URL |
| [19] | 肖清铁, 王经源, 郑新宇, 等. 水稻根系响应镉胁迫的蛋白质差异表达[J]. 生态学报, 2015, 35(24): 8276-8283. |
| Xiao QT, Wang JY, Zheng XY, et al. Analysis of the differently expressed proteins in rice roots in response to cadmium stress[J]. Acta Ecol Sin, 2015, 35(24): 8276-8283. | |
| [20] |
杨婉莹, 孙莎莎, 巩彪, 等. 超表达SlSAMS1对番茄镉胁迫的缓解效应及抗氧化系统的影响[J]. 核农学报, 2020, 34(3): 487-496.
doi: 10.11869/j.issn.100-8551.2020.03.0487 |
| Yang WY, Sun SS, Gong B, et al. Effects of overexpressing SlSAMS1 on tomato tolerance to cadmium toxicity and antioxidant system[J]. J Nucl Agric Sci, 2020, 34(3): 487-496. | |
| [21] |
Chen J, Wang ZB, Liu SD, et al. Nitrogen stress inhibits root growth by regulating cell wall and hormone changes in cotton(Gossypium hirsutum L.)[J]. J Agron Crop Sci, 2021, 207(6): 1006-1023.
doi: 10.1111/jac.v207.6 URL |
| [22] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [23] |
Boerjan W, Bauw G, van Montagu M, et al. Distinct phenotypes generated by overexpression and suppression of S-adenosyl-L-methionine synthetase reveal developmental patterns of gene silencing in tobacco[J]. Plant Cell, 1994, 6(10): 1401-1414.
doi: 10.1105/tpc.6.10.1401 pmid: 7994174 |
| [24] | Heidari P, Mazloomi F, Nussbaumer T, et al. Insights into the SAM synthetase gene family and its roles in tomato seedlings under abiotic stresses and hormone treatments[J]. Plants(Basel), 2020, 9(5): 586. |
| [25] |
赖春旺, 周小娟, 陈燕, 等. 龙眼乙烯合成途径基因鉴定及响应ACC处理的分析[J]. 中国农业科学, 2022, 55(3): 558-574.
doi: 10.3864/j.issn.0578-1752.2022.03.011 |
|
Lai CW, Zhou XJ, Chen Y, et al. Identification of ethylene synthesis pathway genes in longan and its response to ACC treatment[J]. Sci Agric Sin, 2022, 55(3): 558-574.
doi: 10.3864/j.issn.0578-1752.2022.03.011 |
|
| [26] |
Markham GD, Pajares MA. Structure-function relationships in methionine adenosyltransferases[J]. Cell Mol Life Sci, 2009, 66(4): 636-648.
doi: 10.1007/s00018-008-8516-1 pmid: 18953685 |
| [27] | 张寒冰, 张书发, 李毛, 等. 大麦S-腺苷甲硫氨酸合成酶基因HvSAMS2对非生物胁迫响应的表达分析[J]. 农业生物技术学报, 2021, 29(1): 35-46. |
| Zhang HB, Zhang SF, Li M, et al. Expression analysis of S-adenosylmethionine synthetase gene HvSAMS2 from Hordeum vulgare in response to abiotic stress[J]. J Agric Biotechnol, 2021, 29(1): 35-46. | |
| [28] |
谭政委, 李磊, 余永亮, 等. 红花S-腺苷甲硫氨酸合成酶基因的克隆与表达分析[J]. 核农学报, 2021, 35(9): 1994-2001.
doi: 10.11869/j.issn.100-8551.2021.09.1994 |
| Tan ZW, Li L, Yu YL, et al. Cloning and expression analysis of S-adenosylmethionine synthetase gene from Carthamus tinctorius L[J]. J Nucl Agric Sci, 2021, 35(9): 1994-2001. | |
| [29] |
He MW, Wang Y, Wu JQ, et al. Isolation and characterization of S-adenosylmethionine synthase gene from cucumber and responsive to abiotic stress[J]. Plant Physiol Biochem, 2019, 141: 431-445.
doi: 10.1016/j.plaphy.2019.06.006 URL |
| [30] | 何美文. 黄瓜S-腺苷甲硫氨酸合成酶基因鉴定及其在响应盐胁迫中的功能[D]. 南京: 南京农业大学, 2019. |
| He MW. Identification of S-adenosylmethionine synthetase gene and its salt stress response function in cucumber[D]. Nanjing: Nanjing Agricultural University, 2019. | |
| [31] |
Thomas J, Bowman MJ, Vega A, et al. Comparative transcriptome analysis provides key insights into gene expression pattern during the formation of nodule-like structures in Brachypodium[J]. Funct Integr Genomics, 2018, 18(3): 315-326.
doi: 10.1007/s10142-018-0594-z |
| [32] |
Budak H, Zhang BH. microRNAs in model and complex organisms[J]. Funct Integr Genomics, 2017, 17(2/3): 121-124.
doi: 10.1007/s10142-017-0544-1 URL |
| [33] |
Peleman J, Boerjan W, Engler G, et al. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase[J]. Plant Cell, 1989, 1(1): 81-93.
doi: 10.1105/tpc.1.1.81 pmid: 2535470 |
| [34] |
Chen Y, Zou T, McCormick S. S-adenosylmethionine synthetase 3 is important for pollen tube growth[J]. Plant Physiol, 2016, 172(1): 244-253.
doi: 10.1104/pp.16.00774 pmid: 27482079 |
| [35] |
Bai ZT, Qi TX, Liu YC, et al. Alteration of S-adenosylhomocysteine levels affects lignin biosynthesis in switchgrass[J]. Plant Biotechnol J, 2018, 16(12): 2016-2026.
doi: 10.1111/pbi.12935 pmid: 29704888 |
| [36] |
Villalobos DP, Díaz-Moreno SM, Said ESS, et al. Reprogramming of gene expression during compression wood formation in pine: coordinated modulation of S-adenosylmethionine, lignin and lignan related genes[J]. BMC Plant Biol, 2012, 12: 100.
doi: 10.1186/1471-2229-12-100 pmid: 22747794 |
| [37] |
Scully ED, Gries T, Palmer NA, et al. Overexpression of SbMYB60 in Sorghum bicolor impacts both primary and secondary metabolism[J]. New Phytol, 2018, 217(1): 82-104.
doi: 10.1111/nph.14815 pmid: 28944535 |
| [38] |
Srivastava AC, Chen F, Ray T, et al. Loss of function of folylpolyglutamate synthetase 1 reduces lignin content and improves cell wall digestibility in Arabidopsis[J]. Biotechnol Biofuels, 2015, 8: 224.
doi: 10.1186/s13068-015-0403-z pmid: 26697113 |
| [39] |
Murray B, Antonyuk SV, Marina A, et al. Structure and function study of the complex that synthesizes S-adenosylmethionine[J]. IUCrJ, 2014, 1(Pt 4): 240-249.
doi: 10.1107/S2052252514012585 pmid: 25075345 |
| [1] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [2] | 赵光绪, 杨合同, 邵晓波, 崔志豪, 刘红光, 张杰. 一株高效溶磷产红青霉培养条件优化及其溶磷特性[J]. 生物技术通报, 2023, 39(9): 71-83. |
| [3] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [4] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [5] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [6] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [7] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [8] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [9] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [10] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [11] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [12] | 姚晓文, 梁晓, 陈青, 伍春玲, 刘迎, 刘小强, 税军, 乔阳, 毛奕茗, 陈银华, 张银东. 二斑叶螨抗性木薯木质素合成途径基因表达特性研究[J]. 生物技术通报, 2023, 39(2): 161-171. |
| [13] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [14] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [15] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||