








生物技术通报 ›› 2021, Vol. 37 ›› Issue (9): 191-202.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1445
收稿日期:2020-11-25
出版日期:2021-09-26
发布日期:2021-10-25
作者简介:洪军,女,博士,副教授,研究方向:多肽的作用机制;E-mail: 基金资助:
HONG Jun( ), WEI Xia-yi, JI Bing-jie, YE Yan-xin, CHENG Tian-ci
), WEI Xia-yi, JI Bing-jie, YE Yan-xin, CHENG Tian-ci
Received:2020-11-25
Published:2021-09-26
Online:2021-10-25
摘要:
为了探讨铜绿假单胞菌对鲎素抗菌肽的耐药性机制,在转录组水平上通过对铜绿假单胞菌抗鲎素突变株和原始菌株的差异表达基因以及差异表达的sRNA靶基因、SNP的变化进行分析。结果表明,通过GO功能和KEGG通路富集发现差异表达基因与细胞膜的组成部分、核苷酸结合、甲酸脱氢酶(NAD+)活性等功能有关,但无显著富集通路;其中有22个差异表达基因发生SNP的碱基突变,涉及到编码脂质A脱酰基酶、外膜蛋白、冷休克蛋白、以及与脂多糖的修饰相关等已知基因和一些编码的假定蛋白有关。进一步预测与分析找到11个差异表达的sRNA和对应的863个差异表达靶基因,这些sRNA靶基因主要与组氨酸生物合成、高丝氨酸激酶活性、5-羧甲基-2-羟基黏液酸δ-异构酶活性功能最相关。推测铜绿假单胞菌对鲎素的耐药性可能是通过影响氨基酸合成与代谢、膜蛋白的形成与修饰、铁离子代谢等途径来调控的,并使极个别基因发生碱基突变,同时sRNA通过作用于相关的靶基因发挥其调控作用。对于发生SNP突变的基因及sRNA对其靶基因mRNA调控有待进一步验证。
洪军, 卫夏怡, 吉冰洁, 叶延欣, 程天赐. 铜绿假单胞菌对鲎素耐药前后的差异表达基因及SNP变化研究[J]. 生物技术通报, 2021, 37(9): 191-202.
HONG Jun, WEI Xia-yi, JI Bing-jie, YE Yan-xin, CHENG Tian-ci. Change of Differentially Expressed Genes and SNP Before or After Pseudomonas aeruginosa Resistance to Tachyplesin I[J]. Biotechnology Bulletin, 2021, 37(9): 191-202.

图1 PA1.2620 与 PA-99 菌株差异基因的 GO 富集图 横坐标为 -log10(P-value),横坐标越大,表示差异表达基因在该功能注释结果中的富集显著性越可靠。统计学上认为,P-value<0.05 是显著水平,P-value <0.01 是极显著水平。纵坐标为 GO term。下同
Fig. 1 GO enrichment analysis of differentially expressed genes between PA1.2620 vs. PA-99 strains. The abscissa is -log10(P-value). The bigger the value of the abscissa,the more reliable the enrichment significance of differentially expressed genes in the functional annotation results. P-value<0.05 is the significant difference,and P-value<0.01 is the most significant difference. The ordinate is GO term. The same below
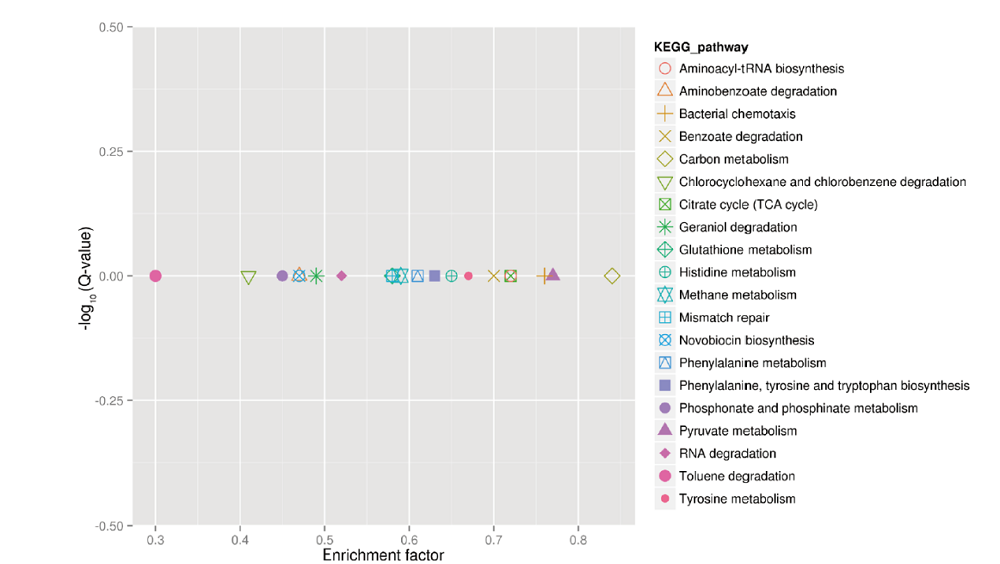
图2 PA1.2620与PA-99菌株差异基因的KEGG富集图 图中每一个图形表示一个KEGG通路,通路名称见右侧图例。横坐标为富集因子(Enrichment factor),表示所有基因中注释到某通路的基因比例与差异基因中注释到该通路的基因比例的比值。富集因子越小,表示差异表达基因在该通路中的富集水平越显著。纵坐标为-log10(Q-value),其中Q-value为多重假设检验校正之后的P-value。因此,纵坐标越大,表示差异表达基因在该通路中的富集显著性越可靠。一般认为,Q-value<0.05是显著水平,Q-value<0.01是极显著水平。下同
Fig.2 KEGG pathway enrichment analysis of differentially expressed genes between PA1.2620 vs. PA-99 strains Each graph in the figure represents one KEGG pathway,whose name is shown in the legend on the right. The abscissa is the enrichment factor,enrichment factor = amount of all genes/amount of DEGs enriched in the pathway in the background gene set. The smaller the enrichment factor,the more significant the enrichment level of differentially expressed genes in this pathway. The ordinate is -log10(Q-value),where Q-value is the P-value after the correction of multiple hypothesis testing. Therefore,the larger the ordinate,the more reliable the enrichment significance of differentially expressed genes in this pathway. It is generally believed that Q-value<0.05 is the significance difference,Q-value<0.01 is the most significant difference. The same below
| PA-99突变株样品 Sample of PA-99 mutant | Number of SNP | Transition | Transversion | Heterozygosity |
|---|---|---|---|---|
| T09 | 139 | 75.54% | 23.74% | 0.72% |
| T10 | 226 | 86.73% | 12.83% | 0.44% |
| T11 | 126 | 72.22% | 26.98% | 0.79% |
表1 SNP位点统计表
Table 1 Statistical table of SNP sites
| PA-99突变株样品 Sample of PA-99 mutant | Number of SNP | Transition | Transversion | Heterozygosity |
|---|---|---|---|---|
| T09 | 139 | 75.54% | 23.74% | 0.72% |
| T10 | 226 | 86.73% | 12.83% | 0.44% |
| T11 | 126 | 72.22% | 26.98% | 0.79% |
| #ID or Gene name | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.15621 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| Novel_244 | — | -1.71631 | Down | 1720641 | 1732256 | 1 | 1732254T>C |
| PA1938 | Hypothetical protein | -1.95428 | Down | 2118926 | 2119747 | 1 | 2119153T>C |
| Novel_497 | — | 1.95302 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.58303 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| Novel_797 | — | -1.77794 | Down | 5343486 | 5344904 | 1 | 5344709G>A |
| Novel_840 | — | -2.52117 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA1428a | Hypothetical protein | 1.07562 | Up | 1552566 | 1554155 | 1 | 1553722G>A |
| PA0193 | Hypothetical protein | 1.186966 | Up | 221585 | 222487 | 1 | 222358C>T |
| exoY | Adenylate cyclase | -1.75024 | Down | 2410344 | 2411480 | 1 | 2411150G>T |
| PA2451 | Hypothetical protein | 1.56984 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| cspD | Cold-shock protein CspD | -1.0514 | Down | 2965201 | 2965473 | 1 | 2965461C>T |
| PA2712 | Hypothetical protein | -1.01673 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
| PA3380 | Hypothetical protein | 2.27038 | Up | 3787021 | 3787479 | 1 | 3787410G>A |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| fepD | Ferric enterobactin transporter FepD | 1.26932 | Up | 4655368 | 4656390 | 1 | 4656097T>C |
| pagL | Lipid A 3-O-deacylase | -2.39699 | Down | 5229459 | 5229980 | 1 | 5229837C>T |
| pmrB | Two-component regulator system Signal sensor kinase PmrB | 1.00016 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| PA4837 | Hypothetical protein | 1.411356 | Up | 5427716 | 5429842 | 1 | 5428274G>A |
| bioD | ATP-dependent dethiobiotin Synthetase BioD | 1.47708 | Up | 563549 | 564235 | 1 | 563798C>T |
| vreR | Sigma factor regulator VreR | 1.68468 | Up | 735487 | 736446 | 1 | 735564C>T |
表2 PA1.2620与PA-99菌株差异表达基因的SNP信息
Table 2 SNP analysis of differentially expressed genes in PA1.2620 vs. PA-99 strains
| #ID or Gene name | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.15621 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| Novel_244 | — | -1.71631 | Down | 1720641 | 1732256 | 1 | 1732254T>C |
| PA1938 | Hypothetical protein | -1.95428 | Down | 2118926 | 2119747 | 1 | 2119153T>C |
| Novel_497 | — | 1.95302 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.58303 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| Novel_797 | — | -1.77794 | Down | 5343486 | 5344904 | 1 | 5344709G>A |
| Novel_840 | — | -2.52117 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA1428a | Hypothetical protein | 1.07562 | Up | 1552566 | 1554155 | 1 | 1553722G>A |
| PA0193 | Hypothetical protein | 1.186966 | Up | 221585 | 222487 | 1 | 222358C>T |
| exoY | Adenylate cyclase | -1.75024 | Down | 2410344 | 2411480 | 1 | 2411150G>T |
| PA2451 | Hypothetical protein | 1.56984 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| cspD | Cold-shock protein CspD | -1.0514 | Down | 2965201 | 2965473 | 1 | 2965461C>T |
| PA2712 | Hypothetical protein | -1.01673 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
| PA3380 | Hypothetical protein | 2.27038 | Up | 3787021 | 3787479 | 1 | 3787410G>A |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| fepD | Ferric enterobactin transporter FepD | 1.26932 | Up | 4655368 | 4656390 | 1 | 4656097T>C |
| pagL | Lipid A 3-O-deacylase | -2.39699 | Down | 5229459 | 5229980 | 1 | 5229837C>T |
| pmrB | Two-component regulator system Signal sensor kinase PmrB | 1.00016 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| PA4837 | Hypothetical protein | 1.411356 | Up | 5427716 | 5429842 | 1 | 5428274G>A |
| bioD | ATP-dependent dethiobiotin Synthetase BioD | 1.47708 | Up | 563549 | 564235 | 1 | 563798C>T |
| vreR | Sigma factor regulator VreR | 1.68468 | Up | 735487 | 736446 | 1 | 735564C>T |
| sRNA ID | Gene length/nt | log2FC | Regulated | sRNA ID | Gene length/nt | log2FC | Regulated |
|---|---|---|---|---|---|---|---|
| Novel_33 | 188 | -3.101050802 | Down | Novel_346 | 136 | -1.138061426 | Down |
| Novel_377 | 378 | -2.065876992 | Down | Novel_593 | 127 | -1.434477351 | Down |
| Novel_583 | 78 | -3.461144761 | Down | Novel_683 | 153 | -1.371013476 | Down |
| Novel_584 | 116 | -2.305895532 | Down | Novel_344 | 60 | 1.764469442 | Up |
| Novel_6 | 94 | -1.576755833 | Down | Novel_5 | 284 | -1.660811503 | Down |
| Novel_853 | 117 | -1.507421364 | Down |
表3 PA1.2620与PA-99菌株差异表达sRNA
Table 3 sRNA of differentially expressed genes in PA1.2620 vs. PA-99 strains
| sRNA ID | Gene length/nt | log2FC | Regulated | sRNA ID | Gene length/nt | log2FC | Regulated |
|---|---|---|---|---|---|---|---|
| Novel_33 | 188 | -3.101050802 | Down | Novel_346 | 136 | -1.138061426 | Down |
| Novel_377 | 378 | -2.065876992 | Down | Novel_593 | 127 | -1.434477351 | Down |
| Novel_583 | 78 | -3.461144761 | Down | Novel_683 | 153 | -1.371013476 | Down |
| Novel_584 | 116 | -2.305895532 | Down | Novel_344 | 60 | 1.764469442 | Up |
| Novel_6 | 94 | -1.576755833 | Down | Novel_5 | 284 | -1.660811503 | Down |
| Novel_853 | 117 | -1.507421364 | Down |
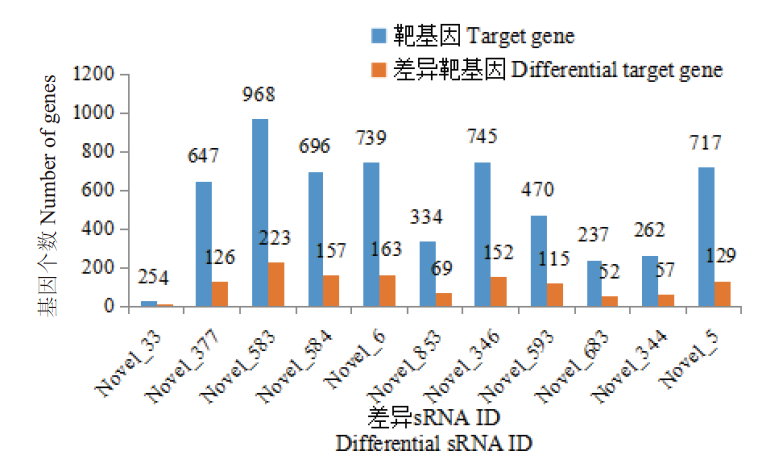
图5 PA1.2620与PA-99菌株差异表达的sRNA靶基因数目统计图
Fig.5 Statistical diagram of the number of sRNA target genes of differentially expressed genes in PA1.2620 vs. PA-99 strains
| Gene ID | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.156212 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| vreR | Sigma factor regulator VreR | 1.684682 | Up | 735487 | 736446 | 1 | 735564C>T |
| Novel_497 | — | 1.953026 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.583033 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| pmrB | Two-component regulator System signal sensor kinase | 1.000160 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| Novel_840 | — | -2.52116 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA2451 | Hypothetical protein | 1.569849 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| PA2712 | Hypothetical protein | -1.01672 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
表4 sRNA相关靶基因的SNP变化分析
Table 4 SNP analysis of sRNA target genes
| Gene ID | Protein name | log2FC | Regulated | Start | End | SNP number | SNP |
|---|---|---|---|---|---|---|---|
| pvdL | Peptide synthase | 1.156212 | Up | 2707666 | 2720694 | 2 | 2712019C>A;2715424T>C |
| vreR | Sigma factor regulator VreR | 1.684682 | Up | 735487 | 736446 | 1 | 735564C>T |
| Novel_497 | — | 1.953026 | Up | 3672989 | 3674050 | 1 | 3673743G>A |
| Novel_745 | — | 2.583033 | Up | 5130955 | 5135726 | 1 | 5130955T>C |
| opr86 | Outer membrane protein Opr86 | -1.51418 | Down | 4085062 | 4087455 | 1 | 4085961C>G |
| cupB3 | Usher CupB3 | -1.59417 | Down | 4565421 | 4567955 | 1 | 4565913A>G |
| pmrB | Two-component regulator System signal sensor kinase | 1.000160 | Up | 5364760 | 5366193 | 1 | 5364817G>T |
| Novel_840 | — | -2.52116 | Down | 5621609 | 5624734 | 1 | 5621617G>A |
| PA2451 | Hypothetical protein | 1.569849 | Up | 2752866 | 2754445 | 1 | 2754392G>A |
| PA2712 | Hypothetical protein | -1.01672 | Down | 3066296 | 3067159 | 1 | 3066971A>G |
| [1] |
Nakamura T, Furunaka H, Miyata T, et al. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab(Tachypleus tridentatus)isolation and chemical structure[J]. Journal of Biological Chemistry, 1988, 263:16709-16713.
pmid: 3141410 |
| [2] |
Anaya-López1 JL, López-Meza JE. Bacterial resistance to cationic antimicrobial peptides[J]. Critical Reviews in Microbiology, 2013, 39(2):180-195.
doi: 10.3109/1040841X.2012.699025 pmid: 22799636 |
| [3] | Cai Y, Chai D, Wang R, et al. Colistin resistance of Acinetobacter baumannii:clinical reports, mechanisms and antimicrobial strategies[J]. Antimicrobial Agents and Chemotherapy, 2012, 67:1607-1615. |
| [4] |
Hong J, Hu JY, Ke F. Experimental induction of bacterial resistance to the antimicrobial peptide tachyplesin I and investigation of the resistance mechanisms[J]. Antimicrobial Agents and Chemotherapy, 2016, 60(10):6067-6075.
doi: 10.1128/AAC.00640-16 pmid: 27480861 |
| [5] | 洪军, 胡建业, 刘坤, 等. 绿脓杆菌对鲎素耐受性特点及耐受性机制[J]. 微生物学报, 2018, 58(9):1593-1604. |
| Hong J, Hu JY, Liu K, et al. Characteristics and resistance of tachyplesin-I resistance in Pseudomonas aeruginosa[J]. Acta Microbiologica Sinica, 2018, 58(9):1593-1604. | |
| [6] | Peng T, Kan J, Lun JS, et al. Identification of novel sRNAs involved in oxidative stress response in the fish pathogen Vibrio alginolyticus by transcriptome analysis[J]. Journal of Fish Diseases, 2019, 42(2):1-15. |
| [7] |
Barnhill EC, Crucello A, Houserova D, et al. Characterization of novel small RNAs(sRNAs)contributing to the desiccation response of Salmonella enterica serovar Typhimurium[J]. RNA Biology, 2019, 16(11):1643-1657.
doi: 10.1080/15476286.2019.1653680 URL |
| [8] | 叶中杨, 邱怀雨, 祝丙华, 等. sRNA调控细菌耐药相关基因表达研究进展[J]. 中国生物工程杂志, 2018, 38(7):89-93. |
| Ye ZY, Qiu HY, Zhu BH, et al. Research progress of sRNA regulates the expression of genes in related with bacterial resistance[J]. China Biotechnology, 2018, 38(7):89-93. | |
| [9] |
Miller CL, Manuel R, Rajasekhar KSL, et al. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions[J]. BMC Microbiology, 2016, 16(1):155.
doi: 10.1186/s12866-016-0771-y URL |
| [10] | 刘翠翠. 福氏志贺菌耐药株的转录组测定及sRNA差异分析[D]. 北京:中国农业科学院, 2014. |
| Liu CC. The transcriptome sequencing analysis and sRNA difference analysis in drug-resistant Shigella flexneri[D]. Beijing:Chinese Academy of Agricultural Sciences, 2014. | |
| [11] |
Zhang YF, Han K, Chandler CE, et al. Probing the sRNA regulatory landscapeof P. aeruginosa:post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility[J]. Molecular Microbiology, 2017, 106(6):919-937.
doi: 10.1111/mmi.2017.106.issue-6 URL |
| [12] | 张栋, 高风英, 卢迈新, 等. 尼罗罗非鱼β2 m基因SNP位点和单倍型与无乳链球菌抗性的关联分析[J]. 水生生物学报, 2018, 42(5):903-912. |
| Zhang D, Gao FY, Lu MX, et al. Association analysis of SNP locus and haplotype of β2m gene in Nile tilapia(Oreochromis niloticus)with its resistance to Streptococcus agalactiae[J]. Acta Hydrobiologica Sinica, 2018, 42(5):903-912. | |
| [13] |
Wang L, Feng Z, Wang X, et al. DEGseq:an R package for identifying differentially expressed genes from RNAseq data[J]. Bioinformatics, 2010, 26:136-138.
doi: 10.1093/bioinformatics/btp612 URL |
| [14] | Ashburner M, Ball CA, Blake JA, et al. Gene ontology:tool for the unification of biology[J]. Nature Genetics, 2000, 25(1):2529. |
| [15] | Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome[J]. Nucleic Acids Research, 2004, 32:D277280. |
| [16] |
Winsor GL, Griffiths EJ, Lo R, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database[J]. Nucleic Acids Research, 2016, 44:D646-D653.
doi: 10.1093/nar/gkv1227 URL |
| [17] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25:402-408.
pmid: 11846609 |
| [18] |
McPhee JB, Bains M, Winsor G, et al. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa[J]. Journal of Bacteriology, 2006, 188:3995-4006.
doi: 10.1128/JB.00053-06 URL |
| [19] |
Moskowitz SM, Brannon MK, Dasgupta N, et al. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients[J]. Antimicrobial Agents and Chemotherapy, 2012, 56(2):1019-1030.
doi: 10.1128/AAC.05829-11 pmid: 22106224 |
| [20] |
Shepherd J, Ibba M. Direction of aminoacylated transfer RNAs into antibiotic synjournal and peptidoglycan-mediated antibiotic resistance[J]. FEBS Letters, 2013, 587(18):2895-2904.
doi: 10.1016/j.febslet.2013.07.036 URL |
| [21] |
Ernst CM, Peschel A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids[J]. Molecular Microbiology, 2011, 80:290-299.
doi: 10.1111/mmi.2011.80.issue-2 URL |
| [22] | 许达, 宗树成, 胡森, 等. 细菌sRNA代谢调控的研究进展[J]. 中国预防兽医学报, 2017, 39(1):80-84. |
| Xu D, Zong SC, Hu S, et al. Research progress on regulation of Bacterial sRNA metabolism[J]. Chinese Journal of Preventive Veterinary Medicine, 2017, 39(1):80-84. |
| [1] | 崔学强, 黄昌艳, 邓杰玲, 李先民, 李秀玲, 张自斌. 基于SLAF-seq技术的石斛兰SNP标记开发及亲缘关系分析[J]. 生物技术通报, 2023, 39(6): 141-148. |
| [2] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [3] | 史建磊, 宰文珊, 苏世闻, 付存念, 熊自立. 番茄青枯病抗性相关miRNA的鉴定与表达分析[J]. 生物技术通报, 2023, 39(5): 233-242. |
| [4] | 肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151. |
| [5] | 王楠楠, 王文佳, 朱强. 植物胁迫相关microRNA研究进展[J]. 生物技术通报, 2022, 38(8): 1-11. |
| [6] | 贺丽娜, 冯源, 石慧敏, 叶建仁. 具有杀线活性马尾松内生细菌的筛选与鉴定[J]. 生物技术通报, 2022, 38(8): 159-166. |
| [7] | 周晓楠, 徐金青, 雷雨晴, 王海庆. 基于GBS-seq的青藏扁蓿豆SNP标记开发[J]. 生物技术通报, 2022, 38(4): 303-310. |
| [8] | 熊和丽, 沙茜, 刘韶娜, 相德才, 张斌, 赵智勇. 单细胞转录组测序技术在动物上的应用研究[J]. 生物技术通报, 2022, 38(3): 226-233. |
| [9] | 胡珊, 梁卫驱, 黄皓, 徐匆, 罗华建, 胡楚维, 黄晓彦, 陈仕丽. 中药渣堆肥中解磷细菌的筛选、鉴定及其拮抗作用[J]. 生物技术通报, 2022, 38(3): 92-102. |
| [10] | 寇航, 王艳梅, 李彤, 薄明井, 张惟材, 熊向华, 黎明. 基于Methylovorus sp. J1-1基因组尺度代谢网络优化吡咯喹啉醌合成[J]. 生物技术通报, 2022, 38(2): 173-183. |
| [11] | 寇佳怡, 王玉玲, 曾睿琳, 兰道亮. 单细胞转录组测序技术及在哺乳动物上的应用[J]. 生物技术通报, 2022, 38(11): 41-48. |
| [12] | 郑青波, 叶娜, 张哓兰, 包鹏甲, 王福彬, 任稳稳, 廖月姣, 阎萍, 潘和平. 天祝白牦牛退行期毛囊细胞亚群鉴定以及特征基因生物信息学分析[J]. 生物技术通报, 2022, 38(10): 262-272. |
| [13] | 张廷焕, 郭宗义, 柴捷, 潘红梅, 张亮, 陈磊, 龙熙. 序列变异对miR-378生物发生以及靶标关系的影响[J]. 生物技术通报, 2022, 38(1): 205-214. |
| [14] | 陈建军, 赵怡迪, 曹香林. 脂多糖对鲤肠上皮细胞转录组模式的调控分析[J]. 生物技术通报, 2021, 37(8): 213-220. |
| [15] | 李玲, 杨丽霞, 郭梅. CNR转录因子在番茄果实成熟过程中的功能[J]. 生物技术通报, 2021, 37(2): 51-62. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||