








生物技术通报 ›› 2022, Vol. 38 ›› Issue (1): 168-178.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0201
张田田( ), 李永臻, 沈国平, 王嵘, 朱德锐, 邢江娃(
), 李永臻, 沈国平, 王嵘, 朱德锐, 邢江娃( )
)
收稿日期:2021-02-03
出版日期:2022-01-26
发布日期:2022-02-22
作者简介:张田田,女,硕士研究生,研究方向:特殊环境微生物学;E-mail: 基金资助:
ZHANG Tian-tian( ), LI Yong-zhen, SHEN Guo-ping, WANG Rong, ZHU De-rui, XING Jiang-wa(
), LI Yong-zhen, SHEN Guo-ping, WANG Rong, ZHU De-rui, XING Jiang-wa( )
)
Received:2021-02-03
Published:2022-01-26
Online:2022-02-22
摘要:
为了解柴达木盆地茶卡盐湖、柯柯盐湖和小柴旦盐湖等三大硫酸镁亚型高盐盐湖可分离嗜盐耐盐菌的种群多样性,采用RM中、高盐培养基筛选分离可培养的嗜盐菌和耐盐菌,扩增16S rRNA基因序列进行种属鉴定和环境因子典范对应分析(CCA),选取优势菌属构建系统发育树,并采用高效液相色谱法(HPLC)检测次级代谢产物四氢嘧啶(Ectoine)积聚量,筛选高产菌株。结果表明三大盐湖共分离获得嗜盐和耐盐菌113株,优势种属为Bacillus(42株)、Staphylococcus(30株)和Halomonas(26株),各丰度与盐湖类型相关;其中中度嗜盐菌居多,其次为耐盐菌和弱嗜盐菌。系统发育分析表明Bacillus属存在5个进化分支,由6个种组成,Halomonas属存在6个进化分支,由7个种组成。HPLC检测四氢嘧啶积聚量,获得具有高产四氢嘧啶潜力菌株7株。这些研究结果说明柴达木盆地硫酸镁亚型高盐盐湖可分离嗜盐耐盐菌的优势菌属以Bacillus、Staphylococcus和Halomonas为主,且多为中度嗜盐菌,其高产四氢嘧啶的潜力菌株可用于后续四氢嘧啶发酵应用研究。
张田田, 李永臻, 沈国平, 王嵘, 朱德锐, 邢江娃. 高盐盐湖可分离嗜盐耐盐菌的种群多样性及四氢嘧啶产量评价[J]. 生物技术通报, 2022, 38(1): 168-178.
ZHANG Tian-tian, LI Yong-zhen, SHEN Guo-ping, WANG Rong, ZHU De-rui, XING Jiang-wa. Population Diversity of Isolated Halophilic and Halotolerant Bacteria from Hypersaline Salt Lakes and Evaluation of Ectoine Production[J]. Biotechnology Bulletin, 2022, 38(1): 168-178.
| 培养基 Culture medium | 分离菌株数量Isolated strain number | 总计 Total | ||
|---|---|---|---|---|
| 茶卡盐湖 Chaka Salt Lake | 柯柯盐湖 Keke Salt Lake | 小柴旦盐湖Xiaochaidan Salt Lake | ||
| 5% NaCl | 38 | 18 | 14 | 70 |
| 10% NaCl | 9 | 22 | 12 | 43 |
表1 分离菌株数据统计
Table 1 Statistics of isolated strains
| 培养基 Culture medium | 分离菌株数量Isolated strain number | 总计 Total | ||
|---|---|---|---|---|
| 茶卡盐湖 Chaka Salt Lake | 柯柯盐湖 Keke Salt Lake | 小柴旦盐湖Xiaochaidan Salt Lake | ||
| 5% NaCl | 38 | 18 | 14 | 70 |
| 10% NaCl | 9 | 22 | 12 | 43 |
| 门Phylum | 属Genus | 5%氯化钠 5% NaCl | 10%氯化钠 10% NaCl | 菌株数量Strain number | 总计Total | ||
|---|---|---|---|---|---|---|---|
| 茶卡盐湖 Chaka Salt Lake | 柯柯盐湖 Keke Salt Lake | 小柴旦盐湖 Xiaochaidan Salt Lake | |||||
| Firmicutes | Bacillus | 32 | 10 | 20(42.6%) | 16(40%) | 6(23.1%) | 42 |
| Staphylococcus | 15 | 15 | 8(17%) | 17(42.5%) | 5(19.3%) | 30 | |
| Enterococcus | 3 | 0 | 0(0%) | 3(7.5%) | 0(0%) | 3 | |
| Planococcus | 1 | 0 | 0(0%) | 0(0%) | 1(3.8%) | 1 | |
| Oceanobacillus | 1 | 0 | 1(2.1%) | 0(0%) | 0(0%) | 1 | |
| Proteobacteria | Halomonas | 11 | 15 | 10(21.3%) | 3(7.5%) | 13(50%) | 26 |
| Pantoea | 6 | 0 | 6(12.8%) | 0(0%) | 0(0%) | 6 | |
| Idiomarina | 1 | 3 | 2(4.2%) | 1(2.5%) | 1(3.8%) | 4 | |
| 总计Total | 70 | 43 | 47 | 40 | 26 | 113 | |
表2 柴达木盆地典型盐湖分离细菌的分子鉴定与丰度统计
Table 2 Molecular identification and abundance statistics of the isolated bacteria from typical salt lakes in Qaidam Basin
| 门Phylum | 属Genus | 5%氯化钠 5% NaCl | 10%氯化钠 10% NaCl | 菌株数量Strain number | 总计Total | ||
|---|---|---|---|---|---|---|---|
| 茶卡盐湖 Chaka Salt Lake | 柯柯盐湖 Keke Salt Lake | 小柴旦盐湖 Xiaochaidan Salt Lake | |||||
| Firmicutes | Bacillus | 32 | 10 | 20(42.6%) | 16(40%) | 6(23.1%) | 42 |
| Staphylococcus | 15 | 15 | 8(17%) | 17(42.5%) | 5(19.3%) | 30 | |
| Enterococcus | 3 | 0 | 0(0%) | 3(7.5%) | 0(0%) | 3 | |
| Planococcus | 1 | 0 | 0(0%) | 0(0%) | 1(3.8%) | 1 | |
| Oceanobacillus | 1 | 0 | 1(2.1%) | 0(0%) | 0(0%) | 1 | |
| Proteobacteria | Halomonas | 11 | 15 | 10(21.3%) | 3(7.5%) | 13(50%) | 26 |
| Pantoea | 6 | 0 | 6(12.8%) | 0(0%) | 0(0%) | 6 | |
| Idiomarina | 1 | 3 | 2(4.2%) | 1(2.5%) | 1(3.8%) | 4 | |
| 总计Total | 70 | 43 | 47 | 40 | 26 | 113 | |
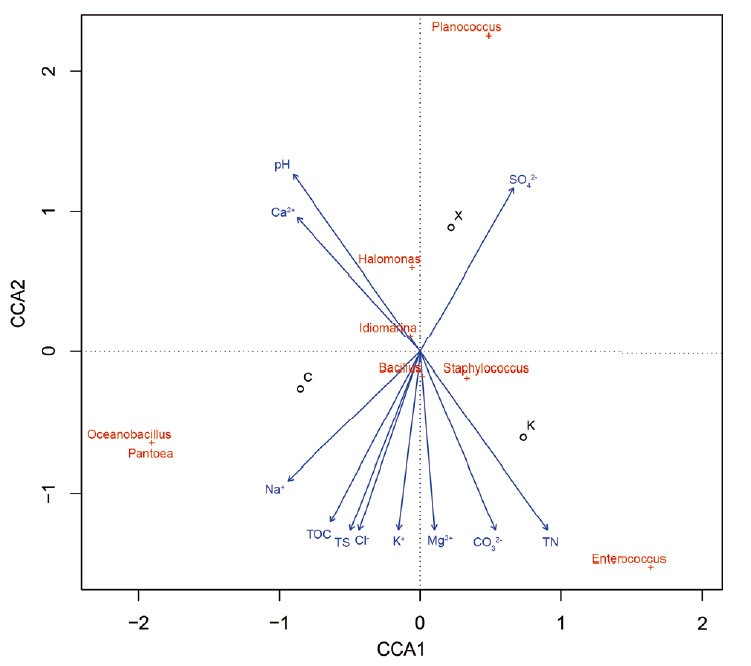
图1 三大盐湖分离细菌环境因子CCA分析 ○ 盐湖;+ 属;→:环境因子
Fig. 1 CCA analysis between environmental factors and bacteria isolated from the three salt lakes ○ Salt lake. + Genus. →:Environmental factor
| Classification of bacteria | Strain code | Salinity range /(mol·L-1) | Optimal salinity /(mol·L-1) | Classification of halophiles | Ectoine accumulation /(mg·g -1CDW) | Strain code | Salinity range /(mol·L-1) | Optimal salinity /(mol·L-1) | Classification of halophiles | Ectoine accumulation /(mg·g -1 CDW) |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus | C10 | 0.5-2.5 | 0.5 | Moderate halophile | 80.08 | K15 | 0-2.5 | 1.5 | Moderate halophile | 147.75 |
| C11 | 0-2.0 | 2.0 | Moderate halophile | 220.56 | K17 | 0.5-2.5 | 1.0 | Moderate halophile | 10 | |
| C12 | 0-3.0 | 0.5 | Moderate halophile | 64.47 | K26 | 0-2.5 | 1.5 | Moderate halophile | ND | |
| C15 | 0.5-3.0 | 0.5 | Moderate halophile | 324.69 | K27 | 0-2.5 | 0.5 | Moderate halophile | 18.20 | |
| C18 | 0-1.5 | 0.5 | Slight halophile | ND | K28 | 0-2.0 | 0.5 | Slight halophile | 32.26 | |
| C20 | 0-1.5 | 0 | Halotolerant | ND | K29 | 0-1.5 | 0.5 | Slight halophile | 30.81 | |
| C22 | 0-1.5 | 0 | Halotolerant | ND | K30 | 0-2.5 | 1.0 | Moderate halophile | 18.50 | |
| C23 | 0-2.0 | 0 | Halotolerant | ND | K31 | 0-2.0 | 0.5 | Slight halophile | 77.60 | |
| C24 | 0-2.5 | 0 | halotolerant | 31.93 | K32 | 0-2.0 | 1.0 | Moderate halophile | 100.35 | |
| C25 | 0-3.0 | 1.0 | Moderate halophile | 80 | K37 | 0-2.5 | 1.5 | Moderate halophile | 135.84 | |
| C26 | 0 -2.0 | 0 | halotolerant | ND | K39 | 0-2.5 | 0.5 | Moderate halophile | 23.88 | |
| C27 | 0-3.0 | 1.0 | Moderate halophile | ND | K40 | 0-3.0 | 0 | Halotolerant | ND | |
| C29 | 0-3.0 | 0 | Halotolerant | ND | K41 | 0-3.0 | 0.5 | Moderate halophile | 10.91 | |
| C30 | 0-2.0 | 0 | Halotolerant | ND | K42 | 0-3.0 | 0 | Halotolerant | ND | |
| C33 | 0-3.0 | 0.5 | Moderate halophile | 50.95 | K48 | 0-3.0 | 0 | Halotolerant | ND | |
| C37 | 0-2.5 | 0 | Halotolerant | ND | X5 | 0.5-3.0 | 1.0 | Moderate halophile | 70.09 | |
| C42 | 0.5-3.0 | 1.0 | Moderate halophile | 65.46 | X10 | 0-2.5 | 0.5 | Moderate halophile | 19.82 | |
| C43 | 0.5-3.0 | 1.5 | Moderate halophile | 133.33 | X11 | 0-2.0 | 0.5 | Slight halophile | 23.99 | |
| C50 | 0-1.5 | 0 | Halotolerant | ND | X20 | 0-2.0 | 0 | Halotolerant | ND | |
| C52 | 0-1.0 | 0 | Halotolerant | ND | X22 | 0-2.5 | 0 | Halotolerant | ND | |
| K11 | 0-2.5 | 1.5 | Moderate halophile | 275.20 | X33 | 0-3.0 | 0 | Halotolerant | ND | |
| Halomonas | C8 | 0-2.5 | 0.5 | Moderate halophile | 30.85 | X4 | 0-3.0 | 1.0 | Moderate halophile | 125.18 |
| C9 | 0-3.0 | 1.5 | Moderate halophile | ND | X6 | 0-3.0 | 1.0 | Moderate halophile | 53.98 | |
| C13 | 0-3.0 | 0.5 | Moderate halophile | 122.38 | X9 | 0-3.0 | 0.5 | Moderate halophile | 41.80 | |
| C17 | 0.5-3.0 | 0.5 | Moderate halophile | 18.66 | X21 | 0-3.0 | 1.0 | Moderate halophile | 100.39 | |
| C34 | 0-3.0 | 0.5 | Moderate halophile | 15.71 | X26 | 0-3.0 | 1.0 | Moderate halophile | 369.10 | |
| C39 | 0-3.0 | 0 | Halotolerant | ND | X27 | 0-3.0 | 1.5 | Moderate halophile | 239.51 | |
| C40 | 0.5-3.0 | 2.0 | Moderate halophile | 11.25 | X28 | 0-3.0 | 1.0 | Moderate halophile | 296.56 | |
| C41 | 0-3.0 | 0.5 | Moderate halophile | 32.35 | X31 | 0-3.0 | 1.5 | Moderate halophile | 41.37 | |
| C44 | 0-3.0 | 2.5 | Moderate halophile | 21.82 | X32 | 0-3.0 | 0.5 | Moderate halophile | 65.23 | |
| C51 | 0-2.5 | 0 | halotolerant | ND | X34 | 0-3.0 | 0.5 | Moderate halophile | 57.61 | |
| K5 | 0-3.0 | 0.5 | Moderate halophile | 9.98 | X35 | 0-3.0 | 1.0 | Moderate halophile | 99.65 | |
| K14 | 0-2.0 | 1.5 | Moderate halophile | 96.81 | X36 | 0-3.0 | 1.0 | Moderate halophile | 208.56 | |
| K38 | 0-3.0 | 0.5 | Moderate halophile | 46.30 | X39 | 0-3.0 | 0.5 | Moderate halophile | 41.26 |
表3 盐湖水体优势菌株生长盐度、最适盐度和四氢嘧啶积聚量
Table 3 Growth salinity,optimal salinity and ectoine accumulation of dominant strains isolated from salt lakes
| Classification of bacteria | Strain code | Salinity range /(mol·L-1) | Optimal salinity /(mol·L-1) | Classification of halophiles | Ectoine accumulation /(mg·g -1CDW) | Strain code | Salinity range /(mol·L-1) | Optimal salinity /(mol·L-1) | Classification of halophiles | Ectoine accumulation /(mg·g -1 CDW) |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus | C10 | 0.5-2.5 | 0.5 | Moderate halophile | 80.08 | K15 | 0-2.5 | 1.5 | Moderate halophile | 147.75 |
| C11 | 0-2.0 | 2.0 | Moderate halophile | 220.56 | K17 | 0.5-2.5 | 1.0 | Moderate halophile | 10 | |
| C12 | 0-3.0 | 0.5 | Moderate halophile | 64.47 | K26 | 0-2.5 | 1.5 | Moderate halophile | ND | |
| C15 | 0.5-3.0 | 0.5 | Moderate halophile | 324.69 | K27 | 0-2.5 | 0.5 | Moderate halophile | 18.20 | |
| C18 | 0-1.5 | 0.5 | Slight halophile | ND | K28 | 0-2.0 | 0.5 | Slight halophile | 32.26 | |
| C20 | 0-1.5 | 0 | Halotolerant | ND | K29 | 0-1.5 | 0.5 | Slight halophile | 30.81 | |
| C22 | 0-1.5 | 0 | Halotolerant | ND | K30 | 0-2.5 | 1.0 | Moderate halophile | 18.50 | |
| C23 | 0-2.0 | 0 | Halotolerant | ND | K31 | 0-2.0 | 0.5 | Slight halophile | 77.60 | |
| C24 | 0-2.5 | 0 | halotolerant | 31.93 | K32 | 0-2.0 | 1.0 | Moderate halophile | 100.35 | |
| C25 | 0-3.0 | 1.0 | Moderate halophile | 80 | K37 | 0-2.5 | 1.5 | Moderate halophile | 135.84 | |
| C26 | 0 -2.0 | 0 | halotolerant | ND | K39 | 0-2.5 | 0.5 | Moderate halophile | 23.88 | |
| C27 | 0-3.0 | 1.0 | Moderate halophile | ND | K40 | 0-3.0 | 0 | Halotolerant | ND | |
| C29 | 0-3.0 | 0 | Halotolerant | ND | K41 | 0-3.0 | 0.5 | Moderate halophile | 10.91 | |
| C30 | 0-2.0 | 0 | Halotolerant | ND | K42 | 0-3.0 | 0 | Halotolerant | ND | |
| C33 | 0-3.0 | 0.5 | Moderate halophile | 50.95 | K48 | 0-3.0 | 0 | Halotolerant | ND | |
| C37 | 0-2.5 | 0 | Halotolerant | ND | X5 | 0.5-3.0 | 1.0 | Moderate halophile | 70.09 | |
| C42 | 0.5-3.0 | 1.0 | Moderate halophile | 65.46 | X10 | 0-2.5 | 0.5 | Moderate halophile | 19.82 | |
| C43 | 0.5-3.0 | 1.5 | Moderate halophile | 133.33 | X11 | 0-2.0 | 0.5 | Slight halophile | 23.99 | |
| C50 | 0-1.5 | 0 | Halotolerant | ND | X20 | 0-2.0 | 0 | Halotolerant | ND | |
| C52 | 0-1.0 | 0 | Halotolerant | ND | X22 | 0-2.5 | 0 | Halotolerant | ND | |
| K11 | 0-2.5 | 1.5 | Moderate halophile | 275.20 | X33 | 0-3.0 | 0 | Halotolerant | ND | |
| Halomonas | C8 | 0-2.5 | 0.5 | Moderate halophile | 30.85 | X4 | 0-3.0 | 1.0 | Moderate halophile | 125.18 |
| C9 | 0-3.0 | 1.5 | Moderate halophile | ND | X6 | 0-3.0 | 1.0 | Moderate halophile | 53.98 | |
| C13 | 0-3.0 | 0.5 | Moderate halophile | 122.38 | X9 | 0-3.0 | 0.5 | Moderate halophile | 41.80 | |
| C17 | 0.5-3.0 | 0.5 | Moderate halophile | 18.66 | X21 | 0-3.0 | 1.0 | Moderate halophile | 100.39 | |
| C34 | 0-3.0 | 0.5 | Moderate halophile | 15.71 | X26 | 0-3.0 | 1.0 | Moderate halophile | 369.10 | |
| C39 | 0-3.0 | 0 | Halotolerant | ND | X27 | 0-3.0 | 1.5 | Moderate halophile | 239.51 | |
| C40 | 0.5-3.0 | 2.0 | Moderate halophile | 11.25 | X28 | 0-3.0 | 1.0 | Moderate halophile | 296.56 | |
| C41 | 0-3.0 | 0.5 | Moderate halophile | 32.35 | X31 | 0-3.0 | 1.5 | Moderate halophile | 41.37 | |
| C44 | 0-3.0 | 2.5 | Moderate halophile | 21.82 | X32 | 0-3.0 | 0.5 | Moderate halophile | 65.23 | |
| C51 | 0-2.5 | 0 | halotolerant | ND | X34 | 0-3.0 | 0.5 | Moderate halophile | 57.61 | |
| K5 | 0-3.0 | 0.5 | Moderate halophile | 9.98 | X35 | 0-3.0 | 1.0 | Moderate halophile | 99.65 | |
| K14 | 0-2.0 | 1.5 | Moderate halophile | 96.81 | X36 | 0-3.0 | 1.0 | Moderate halophile | 208.56 | |
| K38 | 0-3.0 | 0.5 | Moderate halophile | 46.30 | X39 | 0-3.0 | 0.5 | Moderate halophile | 41.26 |
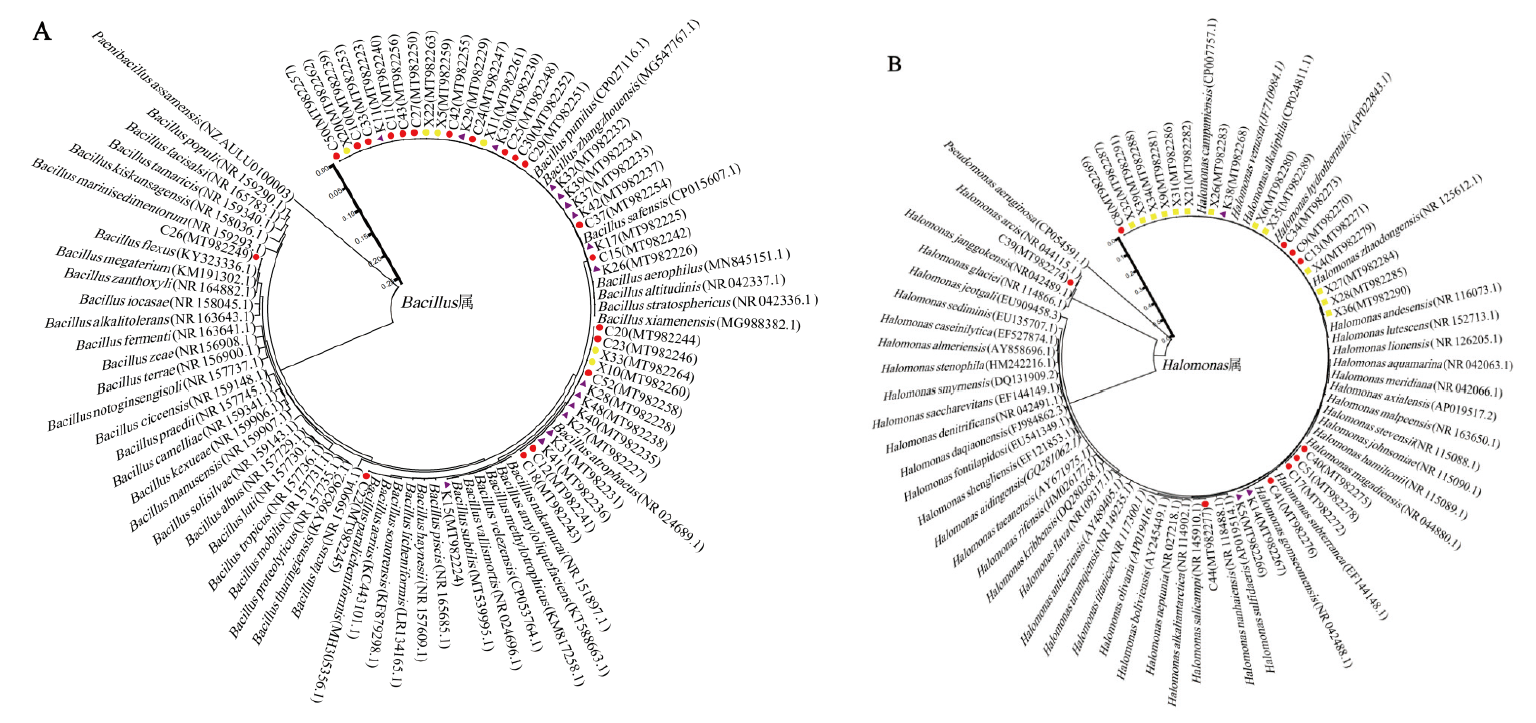
图2 优势菌株系统发育进化树(A:芽孢杆菌属,B:盐单胞菌属) ●茶卡盐湖;▲ 柯柯盐湖;■小柴旦盐湖
Fig. 2 Phylogenetic tree of dominant strains(A:Bacillus. B:Halomonas) ● Chaka Salt Lake. ▲ Keke Salt Lake.■ Xiaochaidan Salt Lake
| [1] | 沈硕. 青藏高原察尔汗盐湖地区可培养中度嗜盐菌的群落结构与多样性[J]. 微生物学报, 2017, 57(4):490-499. |
| Shen S. Community structure and diversity of culturable moderate halophilic bacteria isolated from Qrhan Salt Lake on Qinghai-Tibet Plateau[J]. Acta Microbiol Sin, 2017, 57(4):490-499. | |
| [2] |
Han R, Zhang X, Liu J, et al. Microbial community structure and diversity within hypersaline Keke Salt Lake environments[J]. Can J Microbiol, 2017, 63(11):895-908.
doi: 10.1139/cjm-2016-0773 URL |
| [3] |
Liu W, Zhang GJ, Xian WD, et al. Halomonas xiaochaidanensis sp. nov., isolated from a salt lake sediment[J]. Arch Microbiol, 2016, 198(8):761-766.
doi: 10.1007/s00203-016-1235-3 URL |
| [4] | 郑绵平, 刘喜方. 青藏高原盐湖水化学及其矿物组合特征[J]. 地质学报, 2010, 84(11):1585-1600. |
| Zheng MP, Liu XF. Hydrochemistry and minerals assemblages of salt lakes in the Qinghai-Tibet plateau, China[J]. Acta Geol Sin, 2010, 84(11):1585-1600. | |
| [5] |
Sun YJ, Lai ZP, Long H, et al. Quartz OSL dating of archaeological sites in Xiao Qaidam Lake of the NE Qinghai-Tibetan Plateau and its implications for palaeoenvironmental changes[J]. Quat Geochronol, 2010, 5(2/3):360-364.
doi: 10.1016/j.quageo.2009.02.013 URL |
| [6] | 朱德锐, 韩睿, 石晴, 等. 青藏高原盐湖细菌群落与超盐环境因素的相关性[J]. 中国环境科学, 2017, 37(12):4657-4666. |
| Zhu DR, Han R, Shi Q, et al. The correlation between bacterial communities in salt lakes on the Qinghai-Tibet Plateau and super-salt environmental factors[J]. China Environ Sci, 2017, 37(12):4657-4666. | |
| [7] | 郑绵平, 刘喜方, 赵文. 西藏高原盐湖的构造地球化学和生物学研究[J]. 地质学报, 2007, 81(12):1698-1708. |
| Zheng MP, Liu XF, Zhao W. Tectonogeochemical and biological aspects of salt lakes on the Tibetan Plateau[J]. Acta Geol Sin, 2007, 81(12):1698-1708. | |
| [8] | 李璐, 郝春博, 王丽华, 等. 巴丹吉林沙漠盐湖微生物多样性[J]. 微生物学报, 2015, 55(4):412-424. |
| Li L, Hao CB, Wang LH, et al. Microbial diversity in salt lakes of Badain Jaran Desert[J]. Acta Microbiol Sin, 2015, 55(4):412-424. | |
| [9] | 曹军卫, 沈萍, 李朝阳. 嗜极微生物[M]. 武汉: 武汉大学出版社, 2004. |
| Cao JW, Shen P, Li (C/Z)Y. Extremophiles[M]. Wuhan: Wuhan University Press, 2004. | |
| [10] | Mukherjee P, Mitra A, Roy M. Halomonas rhizobacteria of Avicennia marina of Indian sundarbans promote rice growth under saline and heavy metal stresses through exopolysaccharide production[J]. Front Microbiol, 2019(10):1207. |
| [11] | 周延, 王芳, 王琳. 盐湖开发对柴达木盆地盐湖湖水细菌的多样性影响[J]. 化工进展, 2013, 32(S1):234-239. |
| Zhou Y, Wang F, Wang L. Influence of salt lake exploitation on the bacterial diversity of the salt lakes in the Qaidam Basin[J]. Chem Ind Eng Prog, 2013, 32(S1):234-239. | |
| [12] |
Gan LZ, Li XG, Zhang HM, et al. Preparation, characterization and functional properties of a novel exopolysaccharide produced by the halophilic strain Halomonas saliphila LCB169T[J]. Int J Biol Macromol, 2020, 156:372-380.
doi: 10.1016/j.ijbiomac.2020.04.062 URL |
| [13] |
Wang J, Zhou J, Wang Y, et al. Efficient nitrogen removal in a modified sequencing batch biofilm reactor treating hypersaline mustard Tuber wastewater:The potential multiple pathways and key microorganisms[J]. Water Res, 2020, 177:115734.
doi: 10.1016/j.watres.2020.115734 URL |
| [14] |
Begmatov SA, Selitskaya OV, Vasileva LV, et al. Morphophysiological features of some cultivable bacteria from saline soils of the Aral sea region[J]. Eurasian Soil Sci, 2020, 53(1):90-96.
doi: 10.1134/S1064229320010044 URL |
| [15] |
Wang Q, Cao Z, Liu Q, et al. Enhancement of COD removal in constructed wetlands treating saline wastewater:Intertidal wetland sediment as a novel inoculation[J]. J Environ Manage, 2019, 249:109398.
doi: 10.1016/j.jenvman.2019.109398 URL |
| [16] | 王慧敏, 姚倩倩, 李月月, 等. 渗透压冲击下中度嗜盐菌Halomonas sp. 四氢嘧啶类相容性溶质的合成与释放[J]. 微生物学通报, 2018, 45(4):744-752. |
| Wang HM, Yao QQ, Li YY, et al. Synjournal and release of ectoines in a moderate halophile Halomonas sp. Y subjected to osmotic shocks[J]. Microbiol China, 2018, 45(4):744-752. | |
| [17] |
Nayak PK, Goode M, Chang DP, et al. Ectoine and hydroxyectoine stabilize antibodies in spray-dried formulations at elevated temperature and during a freeze/thaw process[J]. Mol Pharmaceutics, 2020, 17(9):3291-3297.
doi: 10.1021/acs.molpharmaceut.0c00395 URL |
| [18] |
Chookietwattana K, Yuwa-Amornpitak T. Data on soil properties and halophilic bacterial densities in the Na Si Nuan secondary forest at kantharawichai district, maha sarakham, Thailand[J]. Data Brief, 2019, 27:104582.
doi: 10.1016/j.dib.2019.104582 pmid: 31673585 |
| [19] |
Ning Y, Wu X, Zhang C, et al. Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli[J]. Metab Eng, 2016, 36:10-18.
doi: 10.1016/j.ymben.2016.02.013 URL |
| [20] | 赵婉雨, 杨渐, 董海良, 等. 柴达木盆地达布逊盐湖微生物多样性研究[J]. 地球与环境, 2013, 41(4):398-405. |
| Zhao WY, Yang J, Dong HL, et al. Microbial diversity in the hypersaline dabuxun lake in Qaidam basin, China[J]. Earth Environ, 2013, 41(4):398-405. | |
| [21] | 刘文, 杨渐, 吴耿, 等. 青藏高原北部湖泊沉积物中基于不同碳源可培养细菌多样性[J]. 盐湖研究, 2016, 24(2):92-101. |
| Liu W, Yang J, Wu G, et al. Diversity of cultivable bacteria based on different carbon sources in the sediments of lakes on northern Qinghai-Tibet plateau[J]. J Salt Lake Res, 2016, 24(2):92-101. | |
| [22] | 张欣, 刘静, 沈国平, 等. 基于高通量测序研究青藏高原茶卡盐湖微生物多样性[J]. 微生物学通报, 2017, 44(8):1834-1846. |
| Zhang X, Liu J, Shen GP, et al. Illumina-based sequencing analysis of microbial community composition in Chaka Salt Lake in Qinghai-Tibet Plateau[J]. Microbiol China, 2017, 44(8):1834-1846. | |
| [23] | 柴丽红, 崔晓龙, 彭谦, 等. 青海两盐湖细菌多样性研究[J]. 微生物学报, 2004, 44(3):271-275. |
| Chai LH, Cui XL, Peng Q, et al. Bacterial diversity of two salt lakes in Qinghai[J]. Acta Microbiol Sin, 2004, 44(3):271-275. | |
| [24] |
Kumar S, Stecher G, Tamura K. MEGA7:molecular evolutionary genetics analysis version 7. 0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [25] |
Chen YH, Lu CW, Shyu YT, et al. Revealing the saline adaptation strategies of the halophilic bacterium Halomonas beimenensis through high-throughput omics and transposon mutagenesis approaches[J]. Sci Rep, 2017, 7(1):13037.
doi: 10.1038/s41598-017-13450-9 URL |
| [26] | 田磊, 张芳, 沈国平, 等. Ectoine高产菌株Halomonas sp. 的鉴定及紫外诱变选育[J]. 生物学杂志, 2020, 37(4):31-35. |
| Tian L, Zhang F, Shen GP, et al. Identification of high-yielding strain Halomonas sp. XH26 for producing ectoine and UV mutagenesis breeding[J]. J Biol, 2020, 37(4):31-35. | |
| [27] |
Chen J, Liu P, Chu X, et al. Metabolic pathway construction and optimization of Escherichia coli for high-level ectoine production[J]. Curr Microbiol, 2020, 77(8):1412-1418.
doi: 10.1007/s00284-020-01888-6 pmid: 32189048 |
| [28] | 刘静, 张欣, 沈国平, 等. 青藏高原小柴旦盐湖微生物群落结构及多样性[J]. 水生态学杂志, 2017, 38(5):55-64. |
| Liu J, Zhang X, Shen GP, et al. Microbial community structure and diversity of Xiaochaidan salt lake on the Tibetan Plateau[J]. J Hydroecology, 2017, 38(5):55-64. | |
| [29] |
Naghoni A, Emtiazi G, Amoozegar MA, et al. Microbial diversity in the hypersaline Lake Meyghan, Iran[J]. Sci Rep, 2017, 7(1):11522.
doi: 10.1038/s41598-017-11585-3 pmid: 28912589 |
| [30] |
Escudero L, Oetiker N, Gallardo K, et al. A thiotrophic microbial community in an acidic brine lake in Northern Chile[J]. Antonie Van Leeuwenhoek, 2018, 111(8):1403-1419.
doi: 10.1007/s10482-018-1087-8 pmid: 29748902 |
| [31] |
Jacob JH, Hussein EI, Shakhatreh MAK, et al. Microbial community analysis of the hypersaline water of the Dead Sea using high-throughput amplicon sequencing[J]. MicrobiologyOpen, 2017, 6(5):e00500.
doi: 10.1002/mbo3.2017.6.issue-5 URL |
| [32] |
Tazi L, Breakwell DP, Harker AR, et al. Life in extreme environments:microbial diversity in Great Salt Lake, Utah[J]. Extremophiles, 2014, 18(3):525-535.
doi: 10.1007/s00792-014-0637-x URL |
| [33] |
Almeida-Dalmet S, Sikaroodi M, Gillevet P, et al. Temporal study of the microbial diversity of the north arm of great salt lake, Utah, US[J]. Microorganisms, 2015, 3(3):310-326.
doi: 10.3390/microorganisms3030310 pmid: 27682091 |
| [34] |
Boutaiba S, Hacene H, Bidle KA, et al. Microbial diversity of the hypersaline Sidi ameur and himalatt salt lakes of the Algerian Sahara[J]. J Arid Environ, 2011, 75(10):909-916.
pmid: 21909172 |
| [35] |
Jaiani E, Kusradze I, Kokashvili T, et al. Microbial diversity and phage-host interactions in the Georgian coastal area of the black sea revealed by whole genome metagenomic sequencing[J]. Mar Drugs, 2020, 18(11):558.
doi: 10.3390/md18110558 URL |
| [36] | 刘国红, 刘波, 林乃铨, 等. 芽孢杆菌的系统进化及其属分类学特征[J]. 福建农业学报, 2008, 23(4):436-449. |
| Liu GH, Liu B, Lin NQ, et al. Phyletic evolution and taxonomic characteristics of Bacillus[J]. Fujian J Agric Sci, 2008, 23(4):436-449. | |
| [37] | 刘国红, 刘波, 王阶平, 等. 基于可培养方法分析云南腾冲小空山火山谷芽胞杆菌分布特征[J]. 微生物学报, 2019, 59(6):1063-1075. |
| Liu GH, Liu B, Wang JP, et al. Analysis of the distribution characteristics of Bacillus in the volcanic valley of Xiaokong Mountain in Tengchong, Yunnan based on cultivable methods[J]. Acta Microbiol Sin, 2019, 59(6):1063-1075. | |
| [38] |
朱德锐, 刘建, 韩睿, 等. 青海湖嗜盐微生物系统发育与种群多样性[J]. 生物多样性, 2012, 20(4):495-504.
doi: 10.3724/SP.J.1003.2012.10224 |
| Zhu DR, Liu J, Han R, et al. Population diversity and phylogeny of halophiles in the Qinghai Lake[J]. Biodivers Sci, 2012, 20(4):495-504. | |
| [39] |
Neifar M, Chouchane H, Najjari A, et al. Genome analysis provides insights into crude oil degradation and biosurfactant production by extremely halotolerant Halomonas desertis G11 isolated from Chott El-Djerid salt-lake in Tunisian desert[J]. Genomics, 2019, 111(6):1802-1814.
doi: 10.1016/j.ygeno.2018.12.003 URL |
| [40] | 李坤珺, 龙健. 山西运城盐湖嗜盐细菌的系统发育与种群多样性[J]. 贵州农业科学, 2015, 43(11):95-101. |
| Li KJ, Long J. Biodiversity of halophilic BacteriaIn Yuncheng salt lake of Shanxi Province[J]. Guizhou Agric Sci, 2015, 43(11):95-101. | |
| [41] |
León MJ, Hoffmann T, Sánchez-Porro C, et al. Compatible solute synjournal and import by the moderate halophile Spiribacter salinus:physiology and genomics[J]. Front Microbiol, 2018, 9:108.
doi: 10.3389/fmicb.2018.00108 URL |
| [42] |
Chen PW, Cui ZY, Ng HS, et al. Exploring the additive bio-agent impacts upon ectoine production by Halomonas salina DSM5928T using corn steep liquor and soybean hydrolysate as nutrient supplement[J]. J Biosci Bioeng, 2020, 130(2):195-199.
doi: 10.1016/j.jbiosc.2020.03.011 URL |
| [43] |
Krutmann J. Ultraviolet A radiation-induced biological effects in human skin:relevance for photoaging and photodermatosis[J]. J Dermatol Sci, 2000, 23(Suppl 1):S22-S26.
doi: 10.1016/S0923-1811(99)00077-8 URL |
| [44] |
Graf R, Anzali S, Buenger J, et al. The multifunctional role of ectoine as a natural cell protectant[J]. Clin Dermatol, 2008, 26(4):326-333.
doi: 10.1016/j.clindermatol.2008.01.002 URL |
| [45] |
Sydlik U, Gallitz I, Albrecht C, et al. The compatible solute ectoine protects against nanoparticle-induced neutrophilic lung inflammation[J]. Am J Respir Crit Care Med, 2009, 180(1):29-35.
doi: 10.1164/rccm.200812-1911OC URL |
| [46] |
Abdel-Aziz H, Wadie W, Abdallah DM, et al. Novel effects of ectoine, a bacteria-derived natural tetrahydropyrimidine, in experimental colitis[J]. Phytomedicine, 2013, 20(7):585-591.
doi: 10.1016/j.phymed.2013.01.009 pmid: 23453305 |
| [47] |
Thuoc D, Hien TT, Sudesh K. Identification and characterization of ectoine-producing bacteria isolated from Can Gio mangrove soil in Vietnam[J]. Ann Microbiol, 2019, 69(8):819-828.
doi: 10.1007/s13213-019-01474-7 |
| [48] |
Chen WC, Yuan FW, Wang LF, et al. Ectoine production with indigenous Marinococcus sp. MAR2 isolated from the marine environment[J]. Prep Biochem Biotechnol, 2020, 50(1):74-81.
doi: 10.1080/10826068.2019.1663534 URL |
| [49] |
Sajjad W, Qadir S, Ahmad M, et al. Ectoine:a compatible solute in radio-halophilicStenotrophomonassp. WMA-LM19 strain to prevent ultraviolet-induced protein damage[J]. J Appl Microbiol, 2018, 125(2):457-467.
doi: 10.1111/jam.13903 pmid: 29729069 |
| [50] |
Anburajan L, Meena B, Sreelatha T, et al. Ectoine biosynjournal genes from the deep sea halophilic eubacteria, Bacillus clausii NIOT-DSB04:Its molecular and biochemical characterization[J]. Microb Pathog, 2019, 136:103693.
doi: 10.1016/j.micpath.2019.103693 URL |
| [51] |
Chen Q, Zhang LH, Li XL, et al. Poly-β-hydroxybutyrate/ectoine co-production by ectoine-excreting strain Halomonas Salina[J]. Process Biochem, 2014, 49(1):33-37.
doi: 10.1016/j.procbio.2013.09.026 URL |
| [52] |
Zhao Q, Li S, Lv P, et al. High ectoine production by an engineered Halomonas hydrothermalis Y2 in a reduced salinity medium[J]. Microb Cell Fact, 2019, 18(1):184.
doi: 10.1186/s12934-019-1230-x pmid: 31655591 |
| [53] |
Sauer T, Galinski EA. Bacterial milking:a novel bioprocess for production of compatible solutes[J]. Biotechnol Bioeng, 1998, 57(3):306-313.
pmid: 10099207 |
| [54] | 艾丽, 王川, 江科, 等. 古盐井中嗜盐菌的分离及四氢嘧啶的制备[J]. 生物技术, 2018, 28(6):597-602. |
| Ai L, Wang C, Jiang K, et al. Isolation of halophilic bacteria from ancient salt well and production of ectoine[J]. Biotechnology, 2018, 28(6):597-602. | |
| [55] | 姚倩倩, 顾向阳. 中度嗜盐菌Halomonas sp. 的分离和鉴定及其高产四氢嘧啶特性研究[J]. 南京农业大学学报, 2017, 40(1):109-115. |
| Yao QQ, Gu XY. Isolation, identification and the high ectoine-producing characteristics of a moderately halophilic bacterium Halomonas sp. W2[J]. J Nanjing Agric Univ, 2017, 40(1):109-115. |
| [1] | 张淼, 陈裕凤, 陈龙, 黄飘玲, 韦露玲. 不同地区药用植物两面针根际土壤真菌种群多样性差异分析[J]. 生物技术通报, 2020, 36(9): 167-179. |
| [2] | 侯进慧;樊继强;王富威;蔡侃;孔文刚;. 牛蒡根际可培养细菌多样性和镉耐性初步分析[J]. , 2012, 0(08): 158-162. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||