








生物技术通报 ›› 2022, Vol. 38 ›› Issue (12): 100-114.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0346
收稿日期:2022-03-23
出版日期:2022-12-26
发布日期:2022-12-29
作者简介:许博楠,女,硕士研究生,研究方向:合成生物学;E-mail:基金资助:
XU Bo-nan( ), FENG Jia, ZHOU Jian-ting, JIANG Jian-lan(
), FENG Jia, ZHOU Jian-ting, JIANG Jian-lan( )
)
Received:2022-03-23
Published:2022-12-26
Online:2022-12-29
摘要:
无细胞蛋白合成系统(cell-free protein synthesis,CFPS)是一种体外蛋白质合成的高效平台,与体内蛋白合成系统相比,它突破了活细胞的限制,使蛋白合成反应可以在试管中进行,具有灵活、高效、可控性强的特点。近几年来,合成生物学领域新技术的涌现推动了CFPS系统的快速发展,尤其在细胞提取物和模板DNA方面。细胞提取物和模板DNA是CFPS系统中不可或缺的重要组成,很大程度上决定了无细胞反应的效率和成本。本文综述了CFPS系统中细胞提取物和模板DNA方面的研究进展,主要介绍了最近新开发的细胞提取物系统的特点和应用前景,以及质粒DNA模板和线性DNA模板的研究进展。
许博楠, 冯佳, 周见庭, 蒋建兰. 无细胞蛋白合成系统中细胞提取物和模板DNA的研究进展[J]. 生物技术通报, 2022, 38(12): 100-114.
XU Bo-nan, FENG Jia, ZHOU Jian-ting, JIANG Jian-lan. Research Progress of Cell Extracts and Template DNAs in Cell-free Protein Synthesis System[J]. Biotechnology Bulletin, 2022, 38(12): 100-114.
| 细胞提取物系统 System | 优势 Advantages | 劣势 Disadvantages | 应用 Applications |
|---|---|---|---|
| 真核细胞 Eukaryotic | |||
| 小麦胚芽 Wheat germ extract | 无需密码子优化;可高通量合成复杂蛋白质;可合成二硫键桥连的蛋白质;可实现多种蛋白质的正确折叠,使蛋白质具有高溶解性 | 细胞提取物制备费时且成本高;有限的翻译后修饰;缺乏内源性膜结构 | 蛋白质微阵列技术[ |
| 兔网织红细胞 Rabbit reticulocyte extract | 完善且成熟的体系;哺乳动物系统 | 提取物制备需处理活体动物;翻译效率低;需要添加外源性微粒体使蛋白质折叠 | 蛋白质微阵列技术[ |
| 昆虫细胞 Insect cell extract | 细胞易破碎;可进行翻译后修饰;具有内源性微粒体 | 细胞培养成本高;遗传操作少 | 膜蛋白的自动化生产[ |
| 仓鼠卵巢细胞 CHO cell extract | 可进行翻译后修饰;具有内源性微粒体 | 细胞培养成本高 | 生产糖基化蛋白[ |
| 酵母 Yeast extract | 细胞培养快速,提取物制备简单;遗传操作成熟;实现蛋白质的正确折叠;可进行糖基化修饰 | 无类似哺乳动物系统的翻译后修饰;蛋白产量低 | 生产病毒样颗粒用于抗病毒药物研究[ |
| 烟草细胞 Tobacco extract | 细胞提取物制备快速;可进行糖基化修饰和形成二硫键 | 含有内源氨基酸,会影响蛋白质产量 | 研究植物RNA病毒翻译复制机制[ |
| 人类细胞系 Human cell lines | 可进行翻译后修饰;具有内源性微粒体 | 细胞培养成本高;蛋白产量低 | 研究病毒复制机制[ |
| 原核细胞 Prokaryotic | |||
| 大肠杆菌 E. coli extract | 细胞提取物制备简单,成本低;蛋白产量高;遗传操作简便;应用范围广 | 缺乏具有翻译活性的内源性微粒体,有限的翻译后修饰;无法正确折叠复杂蛋白质 | 蛋白的高通量表达[ |
| 枯草芽孢杆菌 B. subtilis extract | 无需密码子优化;遗传操作成熟 | 蛋白质产量低;无翻译后修饰;无内源性微粒体 | 启动子库的原型构建[ |
| 链霉菌 Streptomyces extract | 细胞提取物制备简单 | 无翻译后修饰(未见报道);无内源性微粒体 | 表达高GC含量基因[ |
| 恶臭假单胞菌 P. putida extract | 细胞提取物制备简单 | 目前为止应用有限 | 筛选基因调控元件[ |
| 需钠弧菌 V. natriegens extract | 无需密码子优化;可产生大量有活性的提取物;每个细胞的核糖体浓度高 | 目前为止应用有限 | 启动子库的原型构建[ |
表1 不同细胞提取物系统的特点
Table 1 Features of different cell extract systems
| 细胞提取物系统 System | 优势 Advantages | 劣势 Disadvantages | 应用 Applications |
|---|---|---|---|
| 真核细胞 Eukaryotic | |||
| 小麦胚芽 Wheat germ extract | 无需密码子优化;可高通量合成复杂蛋白质;可合成二硫键桥连的蛋白质;可实现多种蛋白质的正确折叠,使蛋白质具有高溶解性 | 细胞提取物制备费时且成本高;有限的翻译后修饰;缺乏内源性膜结构 | 蛋白质微阵列技术[ |
| 兔网织红细胞 Rabbit reticulocyte extract | 完善且成熟的体系;哺乳动物系统 | 提取物制备需处理活体动物;翻译效率低;需要添加外源性微粒体使蛋白质折叠 | 蛋白质微阵列技术[ |
| 昆虫细胞 Insect cell extract | 细胞易破碎;可进行翻译后修饰;具有内源性微粒体 | 细胞培养成本高;遗传操作少 | 膜蛋白的自动化生产[ |
| 仓鼠卵巢细胞 CHO cell extract | 可进行翻译后修饰;具有内源性微粒体 | 细胞培养成本高 | 生产糖基化蛋白[ |
| 酵母 Yeast extract | 细胞培养快速,提取物制备简单;遗传操作成熟;实现蛋白质的正确折叠;可进行糖基化修饰 | 无类似哺乳动物系统的翻译后修饰;蛋白产量低 | 生产病毒样颗粒用于抗病毒药物研究[ |
| 烟草细胞 Tobacco extract | 细胞提取物制备快速;可进行糖基化修饰和形成二硫键 | 含有内源氨基酸,会影响蛋白质产量 | 研究植物RNA病毒翻译复制机制[ |
| 人类细胞系 Human cell lines | 可进行翻译后修饰;具有内源性微粒体 | 细胞培养成本高;蛋白产量低 | 研究病毒复制机制[ |
| 原核细胞 Prokaryotic | |||
| 大肠杆菌 E. coli extract | 细胞提取物制备简单,成本低;蛋白产量高;遗传操作简便;应用范围广 | 缺乏具有翻译活性的内源性微粒体,有限的翻译后修饰;无法正确折叠复杂蛋白质 | 蛋白的高通量表达[ |
| 枯草芽孢杆菌 B. subtilis extract | 无需密码子优化;遗传操作成熟 | 蛋白质产量低;无翻译后修饰;无内源性微粒体 | 启动子库的原型构建[ |
| 链霉菌 Streptomyces extract | 细胞提取物制备简单 | 无翻译后修饰(未见报道);无内源性微粒体 | 表达高GC含量基因[ |
| 恶臭假单胞菌 P. putida extract | 细胞提取物制备简单 | 目前为止应用有限 | 筛选基因调控元件[ |
| 需钠弧菌 V. natriegens extract | 无需密码子优化;可产生大量有活性的提取物;每个细胞的核糖体浓度高 | 目前为止应用有限 | 启动子库的原型构建[ |
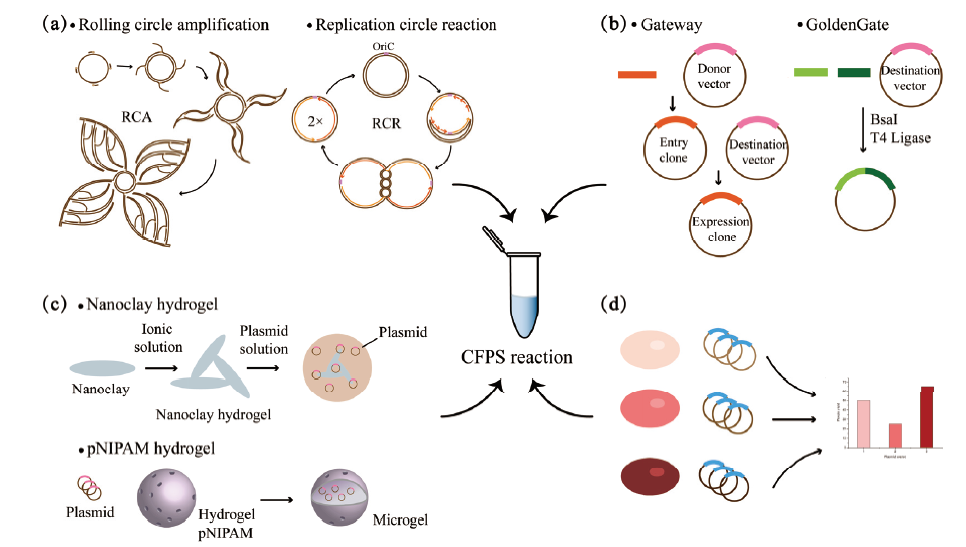
图1 质粒DNA模板在CFPS系统中的研究进展 a:利用质粒DNA体外扩增技术制备大量高纯度DNA,包括RCA与RCR;b:利用高效的分子克隆技术快速制备质粒DNA,包括Gateway克隆系统和GoldenGate克隆系统;c:将质粒DNA与水凝胶结合,如纳米黏土水凝胶和聚N-异丙基丙烯酰胺(pNIPAM)水凝胶,提高DNA局部浓度的同时还可以防止被核酸酶消化;d:质粒DNA来源会影响无细胞蛋白表达水平,应选择与所应用的CFPS系统具有相同细胞背景的表达质粒
Fig. 1 Research progress of plasmid DNA template in CFPS system a:Preparation of large quantities of high-purity DNA by in vitro DNA amplification techniques such as RCA and RCR. b:Rapid preparation of plasmid DNA using efficient cloning techniques,including Gateway cloning system and GoldenGate cloning system. c:Incorporation of plasmids in hydrogels,such as nanoclay hydrogels and poly-N-isopropylacrylamide(pNIPAM)hydrogels,increases the local concentration of DNA while preventing nuclease digestion. d:Plasmid source will affect the level of cell-free protein expression,and an expression plasmid with the same cellular background as the applied CFPS system should be selected
| 质粒DNA的制备策略 Strategies for preparing plasmid DNA | 特点 Characteristic | 参考文献 Reference | |
|---|---|---|---|
| 体外扩增 In vitro amplification | 滚环扩增RCA | 操作简便、特异性强;成本低 | [ |
| 复制周期反应RCR | 短时间内获取大量DNA;与从大肠杆菌中获得的质粒相同 | [ | |
| 高效克隆 Efficient cloning | Gateway | 省时,有助于蛋白高通量表达 | [ |
| GoldenGate | DNA组装工具包/试剂盒,加快蛋白功能表征和路径优化 | [ | |
| 与水凝胶结合 Binding with hydrogel | 黏土水凝胶;基因/黏土/磁性纳米颗粒微凝胶 | 制备简便;DNA局部浓度增加使蛋白产量提高;保护DNA不被核酸酶消化;可以实现蛋白的重复生产 | [ |
| pNIPAM水凝胶 | DNA局部浓度增加使蛋白产量提高;转录和翻译过程局限于表面区域,使mRNA水平下降缓慢和蛋白质翻译迅速 | [ | |
| 质粒DNA来源 Plasmid DNA source | —— | 选择与所应用的CFPS系统具有相同细胞来源的表达质粒会提高蛋白产量 | [ |
表2 质粒DNA模板在CFPS系统中的制备策略
Table 2 Plasmid DNA template preparation strategies in CFPS system
| 质粒DNA的制备策略 Strategies for preparing plasmid DNA | 特点 Characteristic | 参考文献 Reference | |
|---|---|---|---|
| 体外扩增 In vitro amplification | 滚环扩增RCA | 操作简便、特异性强;成本低 | [ |
| 复制周期反应RCR | 短时间内获取大量DNA;与从大肠杆菌中获得的质粒相同 | [ | |
| 高效克隆 Efficient cloning | Gateway | 省时,有助于蛋白高通量表达 | [ |
| GoldenGate | DNA组装工具包/试剂盒,加快蛋白功能表征和路径优化 | [ | |
| 与水凝胶结合 Binding with hydrogel | 黏土水凝胶;基因/黏土/磁性纳米颗粒微凝胶 | 制备简便;DNA局部浓度增加使蛋白产量提高;保护DNA不被核酸酶消化;可以实现蛋白的重复生产 | [ |
| pNIPAM水凝胶 | DNA局部浓度增加使蛋白产量提高;转录和翻译过程局限于表面区域,使mRNA水平下降缓慢和蛋白质翻译迅速 | [ | |
| 质粒DNA来源 Plasmid DNA source | —— | 选择与所应用的CFPS系统具有相同细胞来源的表达质粒会提高蛋白产量 | [ |
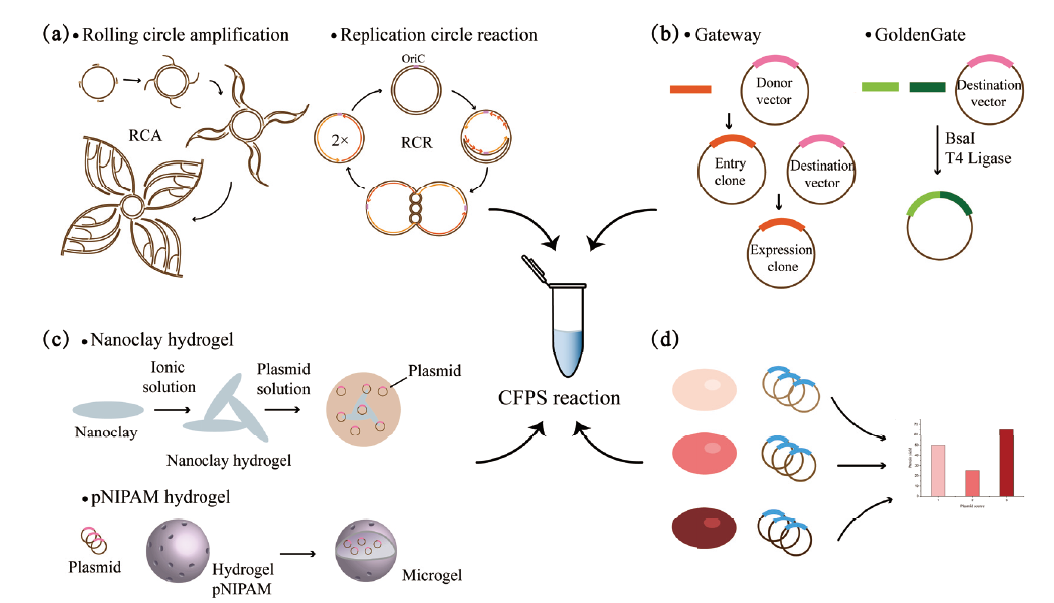
图2 CFPS系统中保护线性DNA模板的研究进展 a:对大肠杆菌基因组进行改造,删除内源性核酸酶基因,制备核酸酶缺陷的细胞提取物用于CFPS反应;b:在CFPS反应中添加核酸酶抑制剂如GamS、Chi DNA以及小分子抑制剂,通过抑制剂与RecBCD的直接结合来降低核酸酶活性;c:对线性DNA进行修饰与设计,包括在DNA两端添加保护序列、将DNA交联成水凝胶、将线性DNA连接成环;d:在CFPS反应中添加DNA结合蛋白如Ku、scCro、Tus-Ter,通过与线性DNA的直接结合来保护它不被降解
Fig. 2 Research progress of protecting linear DNA template in CFPS system a:The E. coli genome is modified to delete endogenous nuclease genes,and nuclease-deficient cell extracts are prepared for CFPS reactions. b:Nuclease inhibitors such as GamS,Chi DNA,and small molecule inhibitors are added to the CFPS reaction to reduce the nuclease activity through the direct binding to RecBCD. c:Modification and design of linear DNA template,including adding protection sequences at both ends of DNA,cross-linking DNA into hydrogels,and ligating into DNA circle. d:DNA-binding proteins such as Ku,scCro,Tus-Ter are added to the CFPS reaction to protect linear DNA from degradation
| 保护线性DNA模板的策略 Strategies for protecting linear DNA templates | 原理 Principles | 蛋白质产量 Protein yield | ||
|---|---|---|---|---|
| 基因组改造 Genomic modifications | ΔrecCBD∷Plac-red-kan-ΔendA[ | 去除提取物中的核酸酶 | 3-6× | 与野生型菌株制备的提取物相比 |
| 核酸酶抑制剂 Nuclease inhibition | GamS[ | 与RecCBD结合 | 37.6% | 与质粒模板相比 |
| 小分子抑制剂[ | 作用于解旋酶或ATP酶从而抑制核酸酶活性 | 200% | 与未添加核酸酶抑制剂相比 | |
| Chi DNA[ | 与RecCBD结合 | 23% | 与质粒模板相比 | |
| 修饰和设计线性DNA模板 Modifications and design | 添加poly(G)序列; 添加T7终止序列[ | 增强mRNA稳定性 | 92%(poly(G)) 265%(T7终止子) | 与未修饰的线性DNA模板相比 |
| 末端添加保护序列[ | 防止核酸酶与线性DNA末端结合 | 75% | 与质粒模板相比 | |
| 水凝胶[ | 防止被核酸酶降解 | 300% | 与液相体系相比 | |
| 线性DNA成环[ | 防止被核酸酶降解 | 100% | 与质粒模板相比 | |
| DNA结合蛋白 DNA-binding protein | Ku[ | 与线性DNA模板结合 | 4.43×(B. subtilis) 1.58×(C. glutamicum) | 与未添加DNA结合蛋白相比 |
| scCro[ | 与线性DNA模板结合 | 6× 18×(V. natriegens) | 与未添加DNA结合蛋白相比 | |
| Tus-Ter[ | Tus蛋白与带有Ter序列的线性DNA模板结合 | 156% 164%(V. natriegens) | 与质粒模板相比 | |
表3 CFPS系统中稳定线性DNA模板的策略比较
Table 3 Comparison of different linear DNA template protection strategies in CFPS system
| 保护线性DNA模板的策略 Strategies for protecting linear DNA templates | 原理 Principles | 蛋白质产量 Protein yield | ||
|---|---|---|---|---|
| 基因组改造 Genomic modifications | ΔrecCBD∷Plac-red-kan-ΔendA[ | 去除提取物中的核酸酶 | 3-6× | 与野生型菌株制备的提取物相比 |
| 核酸酶抑制剂 Nuclease inhibition | GamS[ | 与RecCBD结合 | 37.6% | 与质粒模板相比 |
| 小分子抑制剂[ | 作用于解旋酶或ATP酶从而抑制核酸酶活性 | 200% | 与未添加核酸酶抑制剂相比 | |
| Chi DNA[ | 与RecCBD结合 | 23% | 与质粒模板相比 | |
| 修饰和设计线性DNA模板 Modifications and design | 添加poly(G)序列; 添加T7终止序列[ | 增强mRNA稳定性 | 92%(poly(G)) 265%(T7终止子) | 与未修饰的线性DNA模板相比 |
| 末端添加保护序列[ | 防止核酸酶与线性DNA末端结合 | 75% | 与质粒模板相比 | |
| 水凝胶[ | 防止被核酸酶降解 | 300% | 与液相体系相比 | |
| 线性DNA成环[ | 防止被核酸酶降解 | 100% | 与质粒模板相比 | |
| DNA结合蛋白 DNA-binding protein | Ku[ | 与线性DNA模板结合 | 4.43×(B. subtilis) 1.58×(C. glutamicum) | 与未添加DNA结合蛋白相比 |
| scCro[ | 与线性DNA模板结合 | 6× 18×(V. natriegens) | 与未添加DNA结合蛋白相比 | |
| Tus-Ter[ | Tus蛋白与带有Ter序列的线性DNA模板结合 | 156% 164%(V. natriegens) | 与质粒模板相比 | |
| [1] |
Nielsen J, Larsson C, van Maris A, et al. Metabolic engineering of yeast for production of fuels and chemicals[J]. Curr Opin Biotechnol, 2013, 24(3):398-404.
doi: 10.1016/j.copbio.2013.03.023 URL |
| [2] |
Kwok R. Five hard truths for synthetic biology[J]. Nature, 2010, 463(7279):288-290.
doi: 10.1038/463288a URL |
| [3] | Ranji A, Wu JC, Bundy BC, et al. Transforming synthetic biology with cell-free systems[M]//Synthetic Biology. Amsterdam:Elsevier, 2013:277-301. |
| [4] |
Silverman AD, Karim AS, Jewett MC. Cell-free gene expression:an expanded repertoire of applications[J]. Nat Rev Genet, 2020, 21(3):151-170.
doi: 10.1038/s41576-019-0186-3 pmid: 31780816 |
| [5] |
Carlson ED, Gan R, Hodgman CE, et al. Cell-free protein synthesis:applications come of age[J]. Biotechnol Adv, 2012, 30(5):1185-1194.
doi: 10.1016/j.biotechadv.2011.09.016 pmid: 22008973 |
| [6] | Lu Y. Cell-free synthetic biology:engineering in an open world[J]. Synth Syst Biotechnol, 2017, 2(1):23-27. |
| [7] |
Borsook H. Protein turnover and incorporation of labeled amino acids into tissue proteins in vivo and in vitro[J]. Physiol Rev, 1950, 30(2):206-219.
pmid: 15424034 |
| [8] |
Winnick T. Studies on the mechanism of protein synthesis in embryonic and tumor tissues. I. Evidence relating to the incorporation of labeled amino acids into protein structure in homogenates[J]. Arch Biochem, 1950, 27(1):65-74.
pmid: 15419774 |
| [9] |
Hoagland MB, Stephenson ML, Scott JF, et al. A soluble ribonucleic acid intermediate in protein synthesis[J]. J Biol Chem, 1958, 231(1):241-257.
pmid: 13538965 |
| [10] |
Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides[J]. Proc Natl Acad Sci USA, 1961, 47(10):1588-1602.
doi: 10.1073/pnas.47.10.1588 URL |
| [11] |
Nathans D, Notani G, Schwartz JH, et al. Biosynthesis of the coat protein of coliphage f2 by E. coli extracts[J]. PNAS, 1962, 48(8):1424-1431.
doi: 10.1073/pnas.48.8.1424 URL |
| [12] | Lederman M, Zubay G. DNA-directed peptide synthesis I. A comparison of T2 and Escherichia coli DNA-directed peptide synthesis in two cell-free systems[J]. Biochim Biophys Acta BBA Nucleic Acids Protein Synth, 1967, 149(1):253-258. |
| [13] |
Zubay G. In vitro synthesis of protein in microbial systems[J]. Annu Rev Genet, 1973, 7:267-287.
pmid: 4593305 |
| [14] |
Spirin AS, Baranov VI, Ryabova LA, et al. A continuous cell-free translation system capable of producing polypeptides in high yield[J]. Science, 1988, 242(4882):1162-1164.
pmid: 3055301 |
| [15] |
Calhoun KA, Swartz JR. Energizing cell-free protein synthesis with glucose metabolism[J]. Biotechnol Bioeng, 2005, 90(5):606-613.
pmid: 15830344 |
| [16] |
Goerke AR, Swartz JR. Development of cell-free protein synthesis platforms for disulfide bonded proteins[J]. Biotechnol Bioeng, 2008, 99(2):351-367.
pmid: 17626291 |
| [17] |
Zawada JF, Yin G, Steiner AR, et al. Microscale to manufacturing scale-up of cell-free cytokine production—a new approach for shortening protein production development timelines[J]. Biotechnol Bioeng, 2011, 108(7):1570-1578.
doi: 10.1002/bit.23103 pmid: 21337337 |
| [18] |
Martin RW, des Soye BJ, Kwon YC, et al. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids[J]. Nat Commun, 2018, 9(1):1203.
doi: 10.1038/s41467-018-03469-5 pmid: 29572528 |
| [19] |
Whittaker JW. Cell-free protein synthesis:the state of the art[J]. Biotechnol Lett, 2013, 35(2):143-152.
doi: 10.1007/s10529-012-1075-4 pmid: 23086573 |
| [20] |
Swartz JR. Expanding biological applications using cell-free metabolic engineering:an overview[J]. Metab Eng, 2018, 50:156-172.
doi: S1096-7176(18)30235-0 pmid: 30367967 |
| [21] |
Dopp BJL, Tamiev DD, Reuel NF. Cell-free supplement mixtures:Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract[J]. Biotechnol Adv, 2019, 37(1):246-258.
doi: 10.1016/j.biotechadv.2018.12.006 URL |
| [22] |
Cole SD, Miklos AE, Chiao AC, et al. Methodologies for preparation of prokaryotic extracts for cell-free expression systems[J]. Synth Syst Biotechnol, 2020, 5(4):252-267.
doi: 10.1016/j.synbio.2020.07.006 pmid: 32775710 |
| [23] |
Kigawa T, Yabuki T, Matsuda N, et al. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression[J]. J Struct Funct Genomics, 2004, 5(1/2):63-68.
doi: 10.1023/B:JSFG.0000029204.57846.7d URL |
| [24] |
Liu DV, Zawada JF, Swartz JR. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis[J]. Biotechnol Prog, 2005, 21(2):460-465.
doi: 10.1021/bp049789y URL |
| [25] |
Kim TW, Keum JW, Oh IS, et al. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system[J]. J Biotechnol, 2006, 126(4):554-561.
doi: 10.1016/j.jbiotec.2006.05.014 URL |
| [26] |
Ezure T, Suzuki T, Higashide S, et al. Cell-free protein synthesis system prepared from insect cells by freeze-thawing[J]. Biotechnol Prog, 2006, 22(6):1570-1577.
doi: 10.1021/bp060110v URL |
| [27] |
Didovyk A, Tonooka T, Tsimring L, et al. Rapid and scalable preparation of bacterial lysates for cell-free gene expression[J]. ACS Synth Biol, 2017, 6(12):2198-2208.
doi: 10.1021/acssynbio.7b00253 pmid: 28795570 |
| [28] |
Shrestha P, Holland TM, Bundy BC. Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead Vortex mixing[J]. BioTechniques, 2012, 53(3):163-174.
doi: 10.2144/0000113924 pmid: 22963478 |
| [29] |
Kwon YC, Jewett MC. High-throughput preparation methods of crude extract for robust cell-free protein synthesis[J]. Sci Rep, 2015, 5:8663.
doi: 10.1038/srep08663 URL |
| [30] |
Fujiwara K, Doi N. Biochemical preparation of cell extract for cell-free protein synthesis without physical disruption[J]. PLoS One, 2016, 11(4):e0154614.
doi: 10.1371/journal.pone.0154614 URL |
| [31] |
Michel-Reydellet N, Calhoun K, Swartz J. Amino acid stabilization for cell-free protein synthesis by modification of the Escherichia coli genome[J]. Metab Eng, 2004, 6(3):197-203.
pmid: 15256209 |
| [32] |
des Soye BJ, Gerbasi VR, Thomas PM, et al. A highly productive, one-pot cell-free protein synthesis platform based on genomically recoded Escherichia coli[J]. Cell Chem Biol, 2019, 26(12):1743-1754. e9.
doi: S2451-9456(19)30353-8 pmid: 31706984 |
| [33] | Dudley QM, Nash CJ, Jewett MC. Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates[J]. Synth Biol(Oxf), 2019, 4(1):ysz003. |
| [34] |
Dudley QM, Karim AS, Nash CJ, et al. Cell-free prototyping of limonene biosynthesis using cell-free protein synthesis[J]. bioRxiv, 2020. DOI:10.1101/2020.04.23.057737.
doi: 10.1101/2020.04.23.057737 URL |
| [35] |
Zemella A, Thoring L, Hoffmeister C, et al. Cell-free protein synthesis:pros and cons of prokaryotic and eukaryotic systems[J]. Chembiochem, 2015, 16(17):2420-2431.
doi: 10.1002/cbic.201500340 pmid: 26478227 |
| [36] |
Sawasaki T, Kamura N, Matsunaga S, et al. Arabidopsis HY5 protein functions as a DNA-binding tag for purification and functional immobilization of proteins on agarose/DNA microplate[J]. FEBS Lett, 2008, 582(2):221-228.
doi: 10.1016/j.febslet.2007.12.004 pmid: 18082144 |
| [37] |
Arumugam TU, Ito D, Takashima E, et al. Application of wheat germ cell-free protein expression system for novel malaria vaccine candidate discovery[J]. Expert Rev Vaccines, 2014, 13(1):75-85.
doi: 10.1586/14760584.2014.861747 pmid: 24308585 |
| [38] |
Afonina ZA, Myasnikov AG, Shirokov VA, et al. Conformation transitions of eukaryotic polyribosomes during multi-round translation[J]. Nucleic Acids Res, 2015, 43(1):618-628.
doi: 10.1093/nar/gku1270 pmid: 25520190 |
| [39] |
Oleinikov AV, Gray MD, Zhao J, et al. Self-assembling protein arrays using electronic semiconductor microchips and in vitro translation[J]. J Proteome Res, 2003, 2(3):313-319.
pmid: 12814270 |
| [40] |
He M, Menges M, Groves MA, et al. Selection of a human anti-progesterone antibody fragment from a transgenic mouse library by ARM ribosome display[J]. J Immunol Methods, 1999, 231(1/2):105-117.
doi: 10.1016/S0022-1759(99)00144-1 URL |
| [41] |
Quast RB, Kortt O, Henkel J, et al. Automated production of functional membrane proteins using eukaryotic cell-free translation systems[J]. J Biotechnol, 2015, 203:45-53.
doi: 10.1016/j.jbiotec.2015.03.015 pmid: 25828454 |
| [42] |
Sachse R, Wüstenhagen D, Šamalíková M, et al. Synthesis of membrane proteins in eukaryotic cell-free systems[J]. Eng Life Sci, 2013, 13(1):39-48.
doi: 10.1002/elsc.201100235 URL |
| [43] |
Gurramkonda C, Rao A, Borhani S, et al. Improving the recombinant human erythropoietin glycosylation using microsome supplementation in CHO cell-free system[J]. Biotechnol Bioeng, 2018, 115(5):1253-1264.
doi: 10.1002/bit.26554 pmid: 29384203 |
| [44] |
Wang X, Zhao L, Zhao KN. An optimized yeast cell-free lysate system for in vitro translation of human virus mRNA[J]. Methods Mol Biol, 2014, 1118:219-230.
doi: 10.1007/978-1-62703-782-2_14 pmid: 24395419 |
| [45] |
Sullivan CJ, Pendleton ED, Sasmor HH, et al. A cell-free expression and purification process for rapid production of protein biologics[J]. Biotechnol J, 2016, 11(2):238-248.
doi: 10.1002/biot.201500214 pmid: 26427345 |
| [46] |
安梦楠, 吴元华. BYL无细胞系统对植物RNA病毒翻译复制机制的研究进展[J]. 生物技术通报, 2017, 33(12):45-50.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-0514 URL |
| An MN, Wu YH. Research progress on BYL cell-free systems in plant RNA virus translation and replication mechanisms[J]. Biotechnol Bull, 2017, 33(12):45-50. | |
| [47] |
Buntru M, Hahnengress N, Croon A, et al. Plant-derived cell-free biofactories for the production of secondary metabolites[J]. Front Plant Sci, 2022, 12:794999.
doi: 10.3389/fpls.2021.794999 URL |
| [48] |
Kobayashi T, Mikami S, Yokoyama S, et al. An improved cell-free system for picornavirus synthesis[J]. J Virol Methods, 2007, 142(1/2):182-188.
doi: 10.1016/j.jviromet.2007.01.026 URL |
| [49] |
Kobayashi T, Nakamura Y, Mikami S, et al. Synthesis of encephalomyocarditis virus in a cell-free system:from DNA to RNA virus in one tube[J]. Biotechnol Lett, 2012, 34(1):67-73.
doi: 10.1007/s10529-011-0744-z pmid: 21952914 |
| [50] |
Goshima N, Kawamura Y, Fukumoto A, et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome,[J]. Nat Methods, 2008, 5(12):1011-1017.
pmid: 19054851 |
| [51] |
Hong SH, Kwon YC, Jewett MC. Non-standard amino acid incorporation into proteins using Escherichia coli cell-free protein synthesis[J]. Front Chem, 2014, 2:34.
doi: 10.3389/fchem.2014.00034 pmid: 24959531 |
| [52] |
Kelwick R, Webb AJ, MacDonald JT, et al. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements[J]. Metab Eng, 2016, 38:370-381.
doi: S1096-7176(16)30150-1 pmid: 27697563 |
| [53] |
Tian RZ, Wang MH, Shi JT, et al. Cell-free synthesis system-assisted pathway bottleneck diagnosis and engineering in Bacillus subtilis[J]. Synth Syst Biotechnol, 2020, 5(3):131-136.
doi: 10.1016/j.synbio.2020.06.006 pmid: 32637666 |
| [54] |
Li J, Wang H, Jewett MC. Expanding the palette of Streptomyces-based cell-free protein synthesis systems with enhanced yields[J]. Biochem Eng J, 2018, 130:29-33.
doi: 10.1016/j.bej.2017.11.013 URL |
| [55] | Moore SJ, Lai HE, Needham H, et al. Streptomyces venezuelae TX-TL - a next generation cell-free synthetic biology tool[J]. Biotechnol J, 2017, 12(4): 2017 Apr;12(4). |
| [56] | Wang H, Li J, Jewett MC. Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements[J]. Synth Biol(Oxf), 2018, 3(1):ysy003. |
| [57] |
Failmezger J, Scholz S, Blombach B, et al. Cell-free protein synthesis from fast-growing Vibrio natriegens[J]. Front Microbiol, 2018, 9:1146.
doi: 10.3389/fmicb.2018.01146 pmid: 29910785 |
| [58] |
Takai K, Sawasaki T, Endo Y. Practical cell-free protein synthesis system using purified wheat embryos[J]. Nat Protoc, 2010, 5(2):227-238.
doi: 10.1038/nprot.2009.207 pmid: 20134421 |
| [59] |
Oliver CL, Boyd CD. In vitro translation of messenger RNA in a rabbit reticulocyte lysate cell-free system[J]. Methods Mol Biol, 1985, 2:145-155.
doi: 10.1385/0-89603-064-4:145 pmid: 21374185 |
| [60] |
Ezure T, Suzuki T, Shikata M, et al. A cell-free protein synthesis system from insect cells[J]. Methods Mol Biol, 2010, 607:31-42.
doi: 10.1007/978-1-60327-331-2_4 pmid: 20204846 |
| [61] |
Thoring L, Dondapati SK, Stech M, et al. High-yield production of “difficult-to-express” proteins in a continuous exchange cell-free system based on CHO cell lysates[J]. Sci Rep, 2017, 7(1):11710.
doi: 10.1038/s41598-017-12188-8 pmid: 28916746 |
| [62] |
Gan R, Jewett MC. A combined cell-free transcription-translation system from Saccharomyces cerevisiae for rapid and robust protein synthe[J]. Biotechnol J, 2014, 9(5):641-651.
doi: 10.1002/biot.201300545 pmid: 24677809 |
| [63] |
Buntru M, Vogel S, Spiegel H, et al. Tobacco BY-2 cell-free lysate:an alternative and highly-productive plant-based in vitro translation system[J]. BMC Biotechnol, 2014, 14:37.
doi: 10.1186/1472-6750-14-37 URL |
| [64] | Yadavalli R, Sam-Yellowe T. HeLa based cell free expression systems for expression of Plasmodium rhoptry proteins[J]. J Vis Exp, 2015(100):e52772. |
| [65] |
Harbers M. Wheat germ systems for cell-free protein expression[J]. FEBS Lett, 2014, 588(17):2762-2773.
doi: 10.1016/j.febslet.2014.05.061 pmid: 24931374 |
| [66] |
Thoring L, Zemella A, Wüstenhagen D, et al. Accelerating the production of druggable targets:eukaryotic cell-free systems come into focus[J]. Methods Protoc, 2019, 2(2):30.
doi: 10.3390/mps2020030 URL |
| [67] |
Failmezger J, Rauter M, Nitschel R, et al. Cell-free protein synthesis from non-growing, stressed Escherichia coli[J]. Sci Rep, 2017, 7(1):16524.
doi: 10.1038/s41598-017-16767-7 pmid: 29184159 |
| [68] |
Gu Y, Xu XH, Wu YK, et al. Advances and prospects of Bacillus subtilis cellular factories:from rational design to industrial applications[J]. Metab Eng, 2018, 50:109-121.
doi: 10.1016/j.ymben.2018.05.006 URL |
| [69] |
Legault-Demare L, Chambliss GH. Natural messenger ribonucleic acid-directed cell-free protein-synthesizing system of Bacillus subtilis[J]. J Bacteriol, 1974, 120(3):1300-1307.
doi: 10.1128/jb.120.3.1300-1307.1974 pmid: 4215797 |
| [70] |
Leventhal JM, Chambliss GH. DNA-directed cell-free protein-synthesizing system of Bacillus subtilis[J]. Biochim Biophys Acta, 1979, 564(1):162-171.
pmid: 93969 |
| [71] |
Zaghloul TI, Doi RH.In vitro expression of a Tn9-derived chloramphenicol acetyltransferase gene fusion by using a Bacillus subtilis system[J]. J Bacteriol, 1987, 169(3):1212-1216.
pmid: 3102458 |
| [72] |
Baltz RH. Gifted microbes for genome mining and natural product discovery[J]. J Ind Microbiol Biotechnol, 2017, 44(4/5):573-588.
doi: 10.1007/s10295-016-1815-x URL |
| [73] |
Bentley SD, Chater KF, Cerdeño-Tárraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2)[J]. Nature, 2002, 417(6885):141-147.
doi: 10.1038/417141a URL |
| [74] |
Li J, Wang H, Kwon YC, et al. Establishing a high yielding Streptomyces-based cell-free protein synthesis system[J]. Biotechnol Bioeng, 2017, 114(6):1343-1353.
doi: 10.1002/bit.26253 pmid: 28112394 |
| [75] |
Xu HL, Liu WQ, Li J. Translation related factors improve the productivity of a Streptomyces-based cell-free protein synthesis system[J]. ACS Synth Biol, 2020, 9(5):1221-1224.
doi: 10.1021/acssynbio.0c00140 URL |
| [76] |
Xu HL, Liu WQ, Li J. A Streptomyces-based cell-free protein synthesis system for high-level protein expression[J]. Methods Mol Biol, 2022, 2433:89-103.
doi: 10.1007/978-1-0716-1998-8_5 pmid: 34985739 |
| [77] |
Moore SJ, Lai HG, Chee SM, et al. A Streptomyces venezuelae cell-free toolkit for synthetic biology[J]. ACS Synth Biol, 2021, 10(2):402-411.
doi: 10.1021/acssynbio.0c00581 URL |
| [78] |
Loeschcke A, Thies S. Pseudomonas putida-a versatile host for the production of natural products[J]. Appl Microbiol Biotechnol, 2015, 99(15):6197-6214.
doi: 10.1007/s00253-015-6745-4 pmid: 26099332 |
| [79] |
Nikel PI, Martínez-García E, de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology[J]. Nat Rev Microbiol, 2014, 12(5):368-379.
doi: 10.1038/nrmicro3253 pmid: 24736795 |
| [80] |
Nikel PI, de Lorenzo V. Pseudomonas putida as a functional chassis for industrial biocatalysis:from native biochemistry to trans-metabolism[J]. Metab Eng, 2018, 50:142-155.
doi: 10.1016/j.ymben.2018.05.005 URL |
| [81] |
Nakashima N, Tamura T. Cell-free protein synthesis using cell extract of Pseudomonas fluorescens and CspA promoter[J]. Biochem Biophys Res Commun, 2004, 319(2):671-676.
doi: 10.1016/j.bbrc.2004.05.034 URL |
| [82] | Yim SS, Johns NI, Park J, et al. Multiplex transcriptional characterizations across diverse bacterial species using cell-free systems[J]. Mol Syst Biol, 2019, 15(8):e8875. |
| [83] |
Weinstock MT, Hesek ED, Wilson CM, et al. Vibrio natriegens as a fast-growing host for molecular biology[J]. Nat Methods, 2016, 13(10):849-851.
doi: 10.1038/nmeth.3970 pmid: 27571549 |
| [84] |
Hoff J, Daniel B, Stukenberg D, et al. Vibrio natriegens:an ultrafast-growing marine bacterium as emerging synthetic biology chassis[J]. Environ Microbiol, 2020, 22(10):4394-4408.
doi: 10.1111/1462-2920.15128 URL |
| [85] |
Xu JQ, Yang S, Yang LR. Vibrio natriegens as a host for rapid biotechnology[J]. Trends Biotechnol, 2022, 40(4):381-384.
doi: 10.1016/j.tibtech.2021.10.007 URL |
| [86] | Aiyar SE, Gaal T, Gourse RL. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens[J]. J Bacteriol, 2002, 184 |
| 5):1349-1358. | |
| [87] |
Wiegand DJ, Lee HH, Ostrov N, et al. Establishing a cell-free Vibrio natriegens expression system[J]. ACS Synth Biol, 2018, 7(10):2475-2479.
doi: 10.1021/acssynbio.8b00222 pmid: 30160938 |
| [88] |
Des Soye BJ, Davidson SR, Weinstock MT, et al. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens[J]. ACS Synth Biol, 2018, 7(9):2245-2255.
doi: 10.1021/acssynbio.8b00252 pmid: 30107122 |
| [89] |
Zhang LY, Lin XM, Wang T, et al. Development and comparison of cell-free protein synthesis systems derived from typical bacterial chassis[J]. Bioresour Bioprocess, 2021, 8(1):58.
doi: 10.1186/s40643-021-00413-2 URL |
| [90] |
Dean FB, Nelson JR, Giesler TL, et al. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification[J]. Genome Res, 2001, 11(6):1095-1099.
pmid: 11381035 |
| [91] |
Nojima T, Kimura H, Fujii T. Cell-free protein synthesis conducted by template DNA with repetitive sequence[J]. Chem Lett, 2008, 37(6):648-649.
doi: 10.1246/cl.2008.648 URL |
| [92] |
Kumar G, Chernaya G. Cell-free protein synthesis using multiply-primed rolling circle amplification products[J]. BioTechniques, 2009, 47(1):637-639.
doi: 10.2144/000113171 pmid: 19594449 |
| [93] |
Su’etsugu M, Takada H, Katayama T, et al. Exponential propagation of large circular DNA by reconstitution of a chromosome-replication cycle[J]. Nucleic Acids Res, 2017, 45(20):11525-11534.
doi: 10.1093/nar/gkx822 pmid: 29036468 |
| [94] | Lai HG, Moore S, Polizzi K, et al. EcoFlex:a multifunctional MoClo kit for E. coli synthetic biology[J]. Methods Mol Biol, 2018, 1772:429-444. |
| [95] | Dudley QM, Cai YM, Kallam K, et al. Biofoundry-assisted expression and characterization of plant proteins[J]. Synth Biol(Oxf), 2021, 6(1):ysab029. |
| [96] |
Jiao Y, Liu Y, Luo D, et al. Microfluidic-assisted fabrication of clay microgels for cell-free protein synthesis[J]. ACS Appl Mater Interfaces, 2018, 10(35):29308-29313.
doi: 10.1021/acsami.8b09324 URL |
| [97] |
Chen XJ, Sun Q, Lu Y. Creating a locally crowded environment with nanoclay hydrogels for cell-free biosynthesis[J]. Soft Matter, 2020, 16(22):5132-5138.
doi: 10.1039/d0sm00636j pmid: 32478769 |
| [98] | Wang C, Geng YH, Sun Q, et al. A sustainable and efficient artificial microgel system:toward creating a configurable synthetic cell[J]. Small, 2020, 16(51):e2002313. |
| [99] |
Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination[J]. Genome Res, 2000, 10(11):1788-1795.
pmid: 11076863 |
| [100] |
Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability[J]. PLoS One, 2008, 3(11):e3647.
doi: 10.1371/journal.pone.0003647 URL |
| [101] | Whitfield CJ, Banks AM, Dura G, et al. Cell-free protein synthesis in hydrogel materials[J]. Chem Commun(Camb), 2020, 56 |
| 52):7108-7111. | |
| [102] |
Park N, Um SH, Funabashi H, et al. A cell-free protein-producing gel[J]. Nat Mater, 2009, 8(5):432-437.
doi: 10.1038/nmat2419 pmid: 19329993 |
| [103] |
Kahn JS, Ruiz RCH, Sureka S, et al. DNA microgels as a platform for cell-free protein expression and display[J]. Biomacromolecules, 2016, 17(6):2019-2026.
doi: 10.1021/acs.biomac.6b00183 pmid: 27112709 |
| [104] |
Thiele J, Ma Y, Foschepoth D, et al. DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation[J]. Lab Chip, 2014, 14(15):2651-2656.
doi: 10.1039/c3lc51427g pmid: 24663810 |
| [105] |
Yang DY, Peng SM, Hartman MR, et al. Enhanced transcription and translation in clay hydrogel and implications for early life evolution[J]. Sci Rep, 2013, 3:3165.
doi: 10.1038/srep03165 pmid: 24196527 |
| [106] |
Miyakoshi M, Nishida H, Shintani M, et al. High-resolution mapping of plasmid transcriptomes in different host bacteria[J]. BMC Genomics, 2009, 10:12.
doi: 10.1186/1471-2164-10-12 pmid: 19134166 |
| [107] |
Sun ZZ, Yeung E, Hayes CA, et al. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system[J]. ACS Synth Biol, 2014, 3(6):387-397.
doi: 10.1021/sb400131a pmid: 24303785 |
| [108] |
McSweeney MA, Styczynski MP. Effective use of linear DNA in cell-free expression systems[J]. Front Bioeng Biotechnol, 2021, 9:715328.
doi: 10.3389/fbioe.2021.715328 URL |
| [109] | Schinn SM, Broadbent A, Bradley WT, et al. Protein synthesis directly from PCR:progress and applications of cell-free protein synthesis with linear DNA[J]. Nat Biotechnol, 2016, 33(4):480-487. |
| [110] | Michel-Reydellet N, Woodrow K, Swartz J. Increasing PCR fragment stability and protein yields in a cell-free system with genetically modified Escherichia coli extracts[J]. J Mol Microbiol Biotechnol, 2005, 9(1):26-34. |
| [111] |
Amundsen SK, Spicer T, Karabulut AC, et al. Small-molecule inhibitors of bacterial AddAB and RecBCD helicase-nuclease DNA repair enzymes[J]. ACS Chem Biol, 2012, 7(5):879-891.
doi: 10.1021/cb300018x pmid: 22443934 |
| [112] |
Marshall R, Maxwell CS, Collins SP, et al. Short DNA containing χ sites enhances DNA stability and gene expression in E. coli cell-free transcription-translation systems[J]. Biotechnol Bioeng, 2017, 114(9):2137-2141.
doi: 10.1002/bit.26333 pmid: 28475211 |
| [113] |
Ahn JH, Chu HS, Kim TW, et al. Cell-free synthesis of recombinant proteins from PCR-amplified genes at a comparable productivity to that of plasmid-based reactions[J]. Biochem Biophys Res Commun, 2005, 338(3):1346-1352.
doi: 10.1016/j.bbrc.2005.10.094 URL |
| [114] |
Chen XJ, Lu Y. In silico design of linear DNA for robust cell-free gene expression[J]. Front Bioeng Biotechnol, 2021, 9:670341.
doi: 10.3389/fbioe.2021.670341 URL |
| [115] |
Dopp JL, Rothstein SM, Mansell TJ, et al. Rapid prototyping of proteins:mail order gene fragments to assayable proteins within 24 hours[J]. Biotechnol Bioeng, 2019, 116(3):667-676.
doi: 10.1002/bit.26912 URL |
| [116] |
Yim SS, Johns NI, Noireaux V, et al. Protecting linear DNA templates in cell-free expression systems from diverse bacteria[J]. ACS Synth Biol, 2020, 9(10):2851-2855.
doi: 10.1021/acssynbio.0c00277 URL |
| [117] |
Zhu B, Gan R, Cabezas MD, et al. Increasing cell-free gene expression yields from linear templates in Escherichia coli and Vibrio natriegens extracts by using DNA-binding proteins[J]. Biotechnol Bioeng, 2020, 117(12):3849-3857.
doi: 10.1002/bit.27538 URL |
| [118] |
Norouzi M, Panfilov S, Pardee K. High-efficiency protection of linear DNA in cell-free extracts from Escherichia coli and Vibrio natriegens[J]. ACS Synth Biol, 2021, 10(7):1615-1624.
doi: 10.1021/acssynbio.1c00110 pmid: 34161082 |
| [119] |
Pratt JM, Boulnois GJ, Darby V, et al. Identification of gene products programmed by restriction endonuclease DNA fragments using an E. coli in vitro system[J]. Nucleic Acids Res, 1981, 9(18):4459-4474.
pmid: 6272207 |
| [120] |
Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment[J]. Microbiol Rev, 1994, 58(3):563-602.
doi: 10.1128/mr.58.3.563-602.1994 pmid: 7968924 |
| [121] |
Yu DG, Ellis HM, Lee EC, et al. An efficient recombination system for chromosome engineering in Escherichia coli[J]. PNAS, 2000, 97(11):5978-5983.
pmid: 10811905 |
| [122] |
Sitaraman K, Esposito D, Klarmann G, et al. A novel cell-free protein synthesis system[J]. J Biotechnol, 2004, 110(3):257-263.
pmid: 15163516 |
| [123] | Shrestha P, Smith MT, Bundy BC. Cell-free unnatural amino acid incorporation with alternative energy systems and linear expression templates[J]. Nat Biotechnol, 2014, 31(1):28-34. |
| [124] |
Chen XJ, Liu YY, Hou JQ, et al. A linear DNA template-based framework for site-specific unnatural amino acid incorporation[J]. Synth Syst Biotechnol, 2021, 6(3):192-199.
doi: 10.1016/j.synbio.2021.07.003 pmid: 34401545 |
| [125] |
Cui JH, Wu D, Sun Q, et al. A PEGDA/DNA hybrid hydrogel for cell-free protein synthesis[J]. Front Chem, 2020, 8:28.
doi: 10.3389/fchem.2020.00028 pmid: 32133338 |
| [126] |
Hadi TM, Nozzi N, Melby JO, et al. Rolling circle amplification of synthetic DNA accelerates biocatalytic determination of enzyme activity relative to conventional methods[J]. Sci Rep, 2020, 10(1):10279.
doi: 10.1038/s41598-020-67307-9 pmid: 32581345 |
| [127] |
Sinha KM, Unciuleac MC, Glickman MS, et al. AdnAB:a new DSB-resecting motor-nuclease from Mycobacteria[J]. Genes Dev, 2009, 23(12):1423-1437.
doi: 10.1101/gad.1805709 URL |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||