








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 186-198.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0822
李勇慧1( ), 鲍星星1, 段一珂2, 赵运霞1, 于相丽1, 陈尧1, 张延召1
), 鲍星星1, 段一珂2, 赵运霞1, 于相丽1, 陈尧1, 张延召1
收稿日期:2023-08-22
出版日期:2024-08-26
发布日期:2024-07-02
通讯作者:
李勇慧作者简介:李勇慧,女,硕士,副教授,研究方向:药用植物分子生物学;E-mail: liyonghui@lynu.edu.cn
基金资助:
LI Yong-hui1( ), BAO Xing-xing1, DUAN Yi-ke2, ZHAO Yun-xia1, YU Xiang-li1, CHEN Yao1, ZHANG Yan-zhao1
), BAO Xing-xing1, DUAN Yi-ke2, ZHAO Yun-xia1, YU Xiang-li1, CHEN Yao1, ZHANG Yan-zhao1
Received:2023-08-22
Published:2024-08-26
Online:2024-07-02
摘要:
【目的】鉴定灵宝杜鹃(Rhododendron henanense subsp. lngbaoense)基因组的碱性亮氨酸拉链转录因子(basic leucine zipper, bZIP)家族成员,研究其非生物胁迫下的表达模式。【方法】鉴定RhlbZIP家族成员,构建系统发育树,进行基因结构、保守基序、顺势作用元件和表达模式分析。采用实时荧光定量PCR(RT-qPCR)技术,分析RhlbZIP基因在非生物胁迫中的表达。【结果】共鉴定出57个RhlbZIP基因,划分为11个亚家族,同一亚族成员有相似的结构和基序;不均匀地分布在13条染色体中,共线性分析发现共发生45次复制事件(3对串联复制,42对片段复制);顺式作用元件分析表明,RhlbZIP家族成员可能响应转录、发育、激素调控、生物与非生物胁迫等生物学过程;GO注释分析表明,RhlbZIP基因家族与转录调节、代谢、生物学过程等相关。转录组结果显示,87.72%的WRKY家族基因在根、茎、叶、花和下胚轴5种组织中均表达,但表达水平存在明显差异。RT-qPCR表达模式表明,RhlbZIP4低温处理下显著上调表达;RhlbZIP24在干旱和ABA处理下显著上调;RhlbZIP16在高盐和MeJA处理下显著上调。【结论】灵宝杜鹃基因组中共鉴定出57个RhlbZIP基因,所选的基因在不同处理下均有表达,其中低温、干旱、高盐、ABA与MeJA胁迫处理中的主效基因分别是RhlbZIP4、RhlbZIP24、RhlbZIP16、RhlbZIP24与RhlbZIP16。
李勇慧, 鲍星星, 段一珂, 赵运霞, 于相丽, 陈尧, 张延召. 灵宝杜鹃bZIP家族全基因组鉴定及表达特征分析[J]. 生物技术通报, 2024, 40(8): 186-198.
LI Yong-hui, BAO Xing-xing, DUAN Yi-ke, ZHAO Yun-xia, YU Xiang-li, CHEN Yao, ZHANG Yan-zhao. Genome-wide Identification and Expression Features Analysis of the bZIP Family in Rhododendron henanense subsp. lingbaoense[J]. Biotechnology Bulletin, 2024, 40(8): 186-198.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| EF1α-F | TGTCATCGATGCTCCTGGAC |
| EF1α-R | TCTCGGGTCTGACCATCCTT |
| RhlbZIP4-F | GCAATTTCGGCTCTGTGAGT |
| RhlbZIP4-R | AAGTGGACGGTGAGATTGGT |
| RhlbZIP7-F | GGCCATGGCATCTCATCAAG |
| RhlbZIP7-R | ACGTTGGGATGTCTCCCATT |
| RhlbZIP41-F | GAAAGCACACACGGCTTACT |
| RhlbZIP41-R | TCGAGATTTCGGCTTCGAGA |
表1 RT-qPCR引物序列
Table 1 Primer sequences for RT-qPCR
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| EF1α-F | TGTCATCGATGCTCCTGGAC |
| EF1α-R | TCTCGGGTCTGACCATCCTT |
| RhlbZIP4-F | GCAATTTCGGCTCTGTGAGT |
| RhlbZIP4-R | AAGTGGACGGTGAGATTGGT |
| RhlbZIP7-F | GGCCATGGCATCTCATCAAG |
| RhlbZIP7-R | ACGTTGGGATGTCTCCCATT |
| RhlbZIP41-F | GAAAGCACACACGGCTTACT |
| RhlbZIP41-R | TCGAGATTTCGGCTTCGAGA |
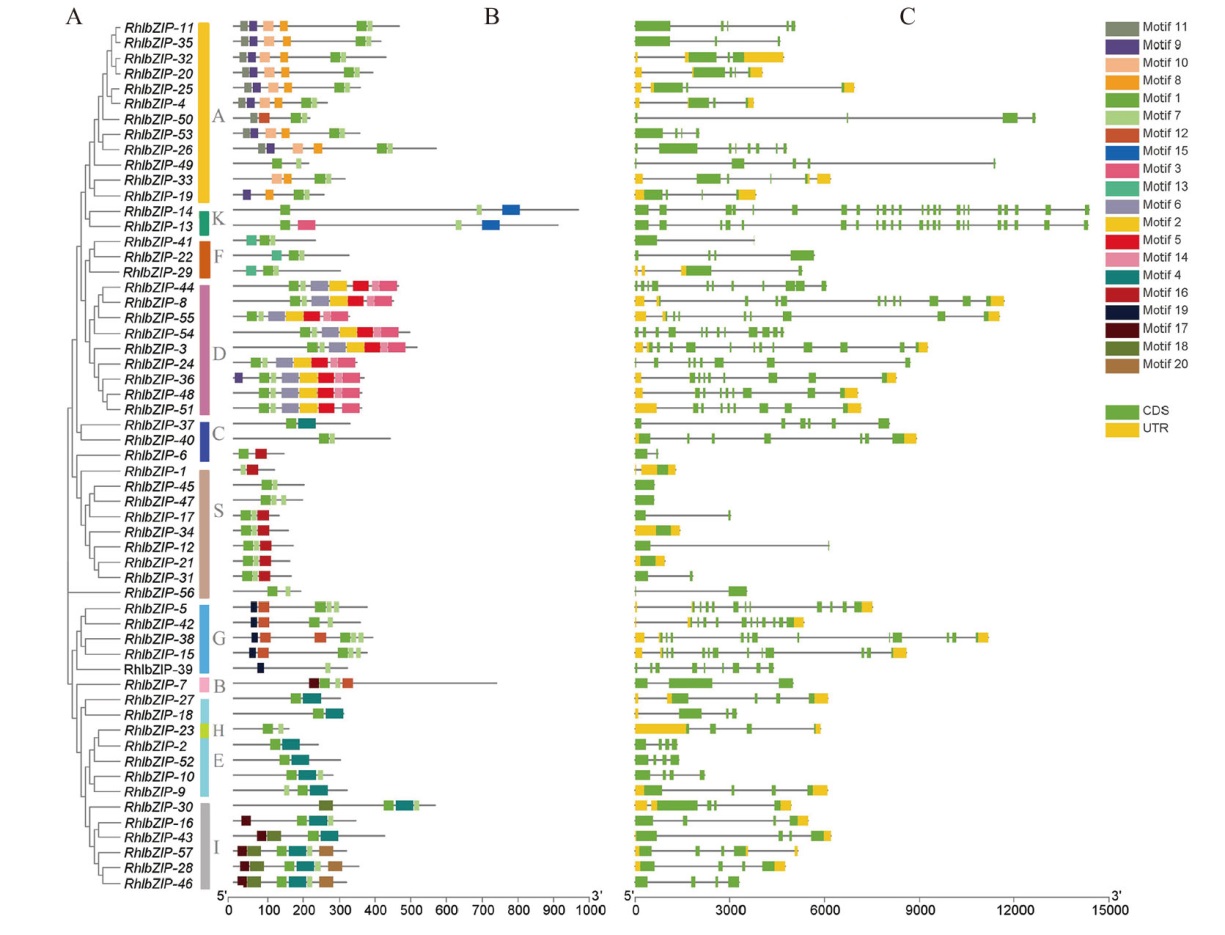
图2 灵宝杜鹃bZIP家族成员系统发育关系(A)、基序组成(B)和基因结构(C)
Fig. 2 Phylogenetic relationship(A), motif composition(B), and gene structure combination(C)of members of the bZIP family of R. henanense subsp. lingbaoense
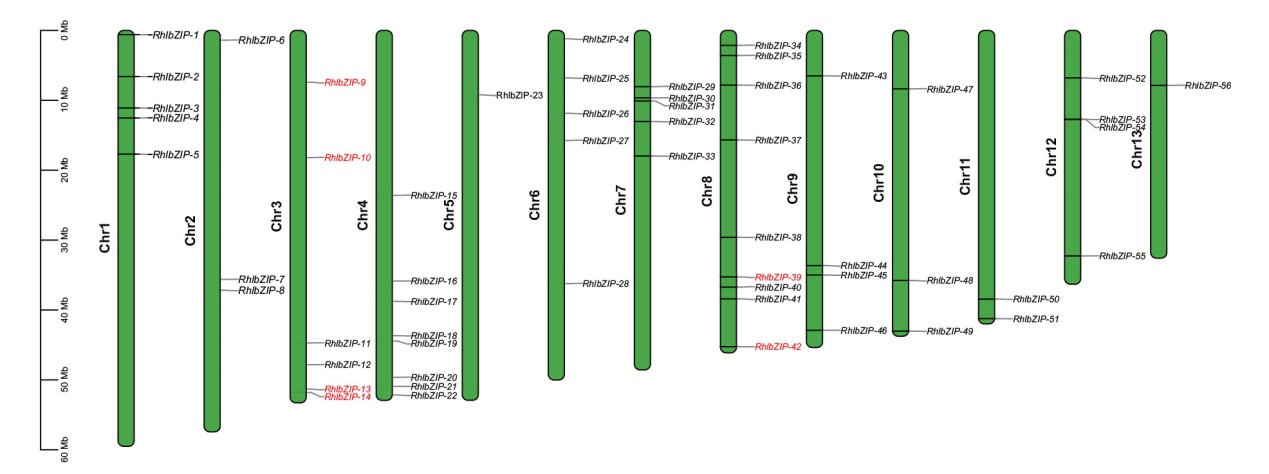
图3 灵宝杜鹃bZIP基因家族染色体定位 红色基因为串联重复基因
Fig. 3 Chromosomal localization of bZIP family genes in R. henanense subsp. Lingbaoense The red genes are repeated in series
| 基因对 Gene pairs | Ka | Ks | Ka/Ks值 |
|---|---|---|---|
| RhlbZIP-2&RhlbZIP-52 | 0.965 8 | 1.110 4 | 0.869 8 |
| RhlbZIP-3&RhlbZIP-54 | 5.273 6 | 2.352 6 | 2.241 6 |
| RhlbZIP-4&RhlbZIP-25 | 2.243 5 | 3.787 2 | 0.592 4 |
| RhlbZIP-7&RhlbZIP-18 | 4.907 7 | 2.147 5 | 2.285 3 |
| RhlbZIP-12&RhlbZIP-21 | 1.765 8 | 3.384 7 | 0.521 7 |
| RhlbZIP-13&RhlbZIP-14 | 1.047 2 | 1.365 0 | 0.767 1 |
| RhlbZIP-15&RhlbZIP-38 | 1.438 8 | 2.038 5 | 0.705 8 |
| RhlbZIP-19&RhlbZIP-33 | 0.970 8 | 1.108 9 | 0.875 5 |
| RhlbZIP-20&RhlbZIP-32 | 1.027 2 | 0.905 2 | 1.134 8 |
| RhlbZIP-26&RhlbZIP-53 | 2.080 5 | 3.975 9 | 0.523 3 |
| RhlbZIP-28&RhlbZIP-46 | 1.047 1 | 0.846 9 | 1.236 5 |
| RhlbZIP-30&RhlbZIP-43 | 0.962 8 | 1.134 1 | 0.849 0 |
| RhlbZIP-31&RhlbZIP-34 | 0.931 3 | 1.280 4 | 0.727 4 |
| RhlbZIP-36&RhlbZIP-48 | 2.289 1 | 4.049 4 | 0.565 3 |
| RhlbZIP-37&RhlbZIP-40 | 1.020 4 | 0.936 5 | 1.089 6 |
| RhlbZIP-44&RhlbZIP-55 | 0.976 2 | 1.078 6 | 0.905 1 |
表2 16个基因对的Ks、Ka和Ka/Ks的比值
Table 2 Ks, Ka and Ka/Ks ratios of the sixteen gene pairs
| 基因对 Gene pairs | Ka | Ks | Ka/Ks值 |
|---|---|---|---|
| RhlbZIP-2&RhlbZIP-52 | 0.965 8 | 1.110 4 | 0.869 8 |
| RhlbZIP-3&RhlbZIP-54 | 5.273 6 | 2.352 6 | 2.241 6 |
| RhlbZIP-4&RhlbZIP-25 | 2.243 5 | 3.787 2 | 0.592 4 |
| RhlbZIP-7&RhlbZIP-18 | 4.907 7 | 2.147 5 | 2.285 3 |
| RhlbZIP-12&RhlbZIP-21 | 1.765 8 | 3.384 7 | 0.521 7 |
| RhlbZIP-13&RhlbZIP-14 | 1.047 2 | 1.365 0 | 0.767 1 |
| RhlbZIP-15&RhlbZIP-38 | 1.438 8 | 2.038 5 | 0.705 8 |
| RhlbZIP-19&RhlbZIP-33 | 0.970 8 | 1.108 9 | 0.875 5 |
| RhlbZIP-20&RhlbZIP-32 | 1.027 2 | 0.905 2 | 1.134 8 |
| RhlbZIP-26&RhlbZIP-53 | 2.080 5 | 3.975 9 | 0.523 3 |
| RhlbZIP-28&RhlbZIP-46 | 1.047 1 | 0.846 9 | 1.236 5 |
| RhlbZIP-30&RhlbZIP-43 | 0.962 8 | 1.134 1 | 0.849 0 |
| RhlbZIP-31&RhlbZIP-34 | 0.931 3 | 1.280 4 | 0.727 4 |
| RhlbZIP-36&RhlbZIP-48 | 2.289 1 | 4.049 4 | 0.565 3 |
| RhlbZIP-37&RhlbZIP-40 | 1.020 4 | 0.936 5 | 1.089 6 |
| RhlbZIP-44&RhlbZIP-55 | 0.976 2 | 1.078 6 | 0.905 1 |
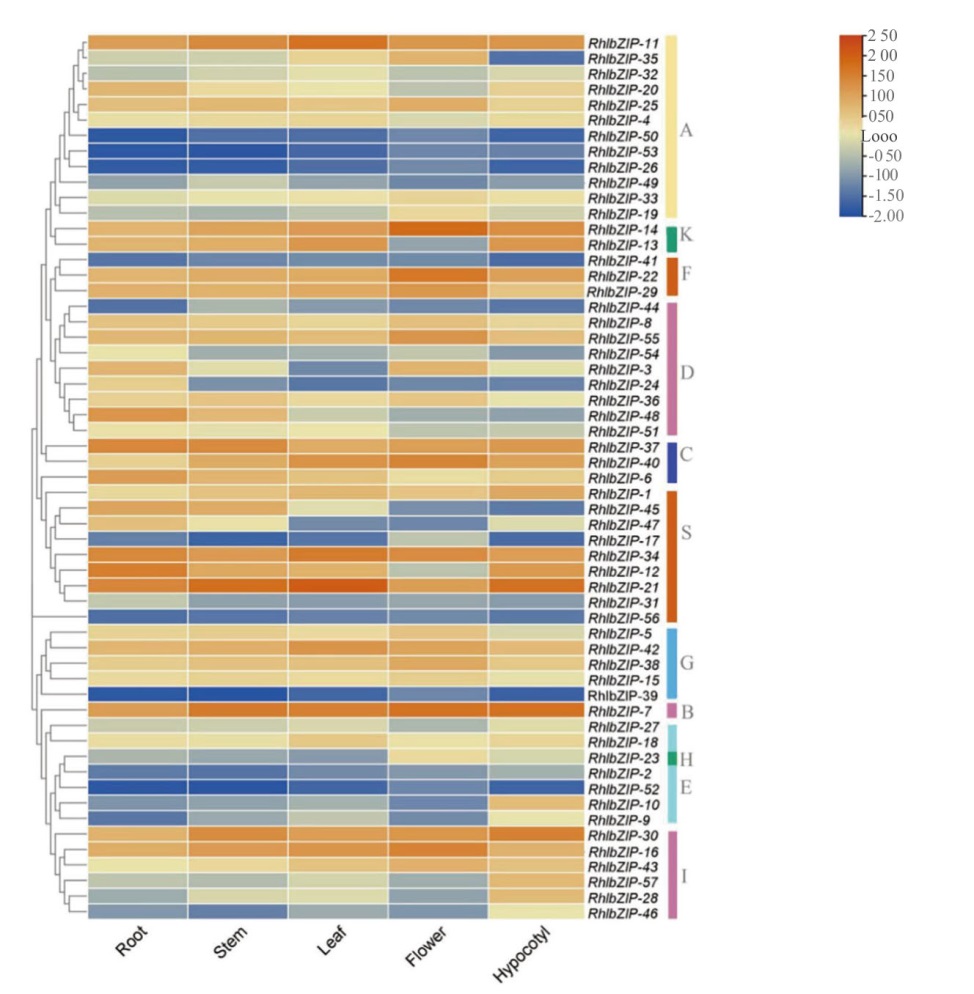
图7 RhlbZIP基因在不同组织(根、茎、叶、花和下胚轴)中表达谱的热图 根据色码显示log10(FPKM)值(右上)
Fig. 7 Hierarchical clustering of expression profiles of RhlbZIP genes in different tissues(root, stem, leaf, flower, and hypocotyl) log10(FPKM)values showed according to the color code(Top right)
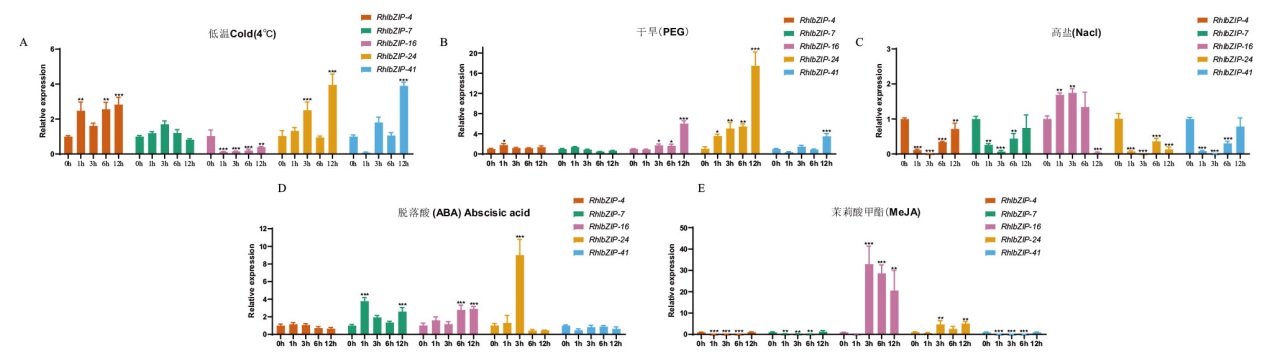
图8 灵宝杜鹃基因在不同处理下表达量分析 与0 h对照相比,* P < 0.05,**P < 0.01,***P<0.001
Fig. 8 Expression analysis of three genes of in R. henanense subsp. lingbaoense under different treatments Compared with 0 h control, * P< 0.05, **P< 0.01, ***P<0. 001
| [1] | 岳伏牛, 韩铁艳, 张亚芳. 灵宝杜鹃种质资源的生境条件调查保护与利用[J]. 特种经济动植物, 2019, 22(11): 22, 25. |
| Yue FN, Han TY, Zhang YF. Investigation, protection and utilization of the habitat conditions of Rhododendron henanense subsp. lingbaoense germplasm resource[J]. Spec Econ Anim Plant, 2019, 22(11): 22, 25. | |
| [2] | 韩军旺, 张莹, 袁志良, 等. 河南小秦岭国家级自然保护区野生灵宝杜鹃的引种及开发[J]. 安徽农业科学, 2008, 36(34): 14954-14955. |
| Han JW, Zhang Y, Yuan ZL, et al. Study on the introduction and development of wild Rhododendron lingbaoense in Xiaoqinling national nature reserve of Henan[J]. J Anhui Agric Sci, 2008, 36(34): 14954-14955. | |
| [3] | 马宏, 李太强, 刘雄芳, 等. 杜鹃属植物保护生物学研究进展[J]. 世界林业研究, 2017, 30(4): 13-17. |
| Ma H, Li TQ, Liu XF, et al. Research progress in conservation biology of Rhododendron[J]. World For Res, 2017, 30(4): 13-17. | |
| [4] | 周晓君, 王海亮, 李方玲, 等. 基于RAD-seq技术开发灵宝杜鹃多态性SSR标记[J]. 农业生物技术学报, 2019, 27(1): 55-62. |
| Zhou XJ, Wang HL, Li FL, et al. Development of polymorphic SSR markers in Rhododendron henanense subsp. lingbaoense based on RAD-seq[J]. J Agric Biotechnol, 2019, 27(1): 55-62. | |
| [5] | 常国栋. 照山白的化学成分及生物活性研究[D]. 保定: 河北大学, 2011. |
| Chang GD. Study on chemical constituents and bioactivities from Rhododendron micranthum turcz[D]. Baoding: Hebei University, 2011. | |
| [6] | Ku SK, Zhou W, Lee W, et al. Anti-inflammatory effects of hyperoside in human endothelial cells and in mice[J]. Inflammation, 2015, 38(2): 784-799. |
| [7] | Li TQ, Liu XF, Li ZH, et al. Study on reproductive biology of Rhododendron longipedicellatum: a newly discovered and special threatened plant surviving in limestone habitat in southeast Yunnan, China[J]. Front Plant Sci, 2018, 9: 33. |
| [8] | Liang JY, You CX, Guo SS, et al. Chemical constituents of the essential oil extracted from Rhododendron thymifolium and their insecticidal activities against Liposcelis bostrychophila or Tribolium castaneum[J]. Ind Crops Prod, 2016, 79: 267-273. |
| [9] |
Zhao J, Guo RR, Guo CL, et al. Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family[J]. Front Plant Sci, 2016, 7: 376.
doi: 10.3389/fpls.2016.00376 pmid: 27066030 |
| [10] |
Nijhawan A, Jain M, Tyagi AK, et al. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice[J]. Plant Physiol, 2008, 146(2): 333-350.
doi: 10.1104/pp.107.112821 pmid: 18065552 |
| [11] | Gai WX, Ma X, Qiao YM, et al. Characterization of the bZIP transcription factor family in pepper(Capsicum annuum L.): CabZIP25 positively modulates the salt tolerance[J]. Front Plant Sci, 2020, 11: 139. |
| [12] |
Zhang Y, Gao WL, Li HT, et al. Genome-wide analysis of the bZIP gene family in Chinese jujube(Ziziphus jujuba Mill.)[J]. BMC Genomics, 2020, 21(1): 483.
doi: 10.1186/s12864-020-06890-7 pmid: 32664853 |
| [13] |
Jakoby M, Weisshaar B, Dröge-Laser W, et al. bZIP transcription factors in Arabidopsis[J]. Trends Plant Sci, 2002, 7(3): 106-111.
doi: 10.1016/s1360-1385(01)02223-3 pmid: 11906833 |
| [14] |
Zhao K, Chen S, Yao WJ, et al. Genome-wide analysis and expression profile of the bZIP gene family in poplar[J]. BMC Plant Biol, 2021, 21(1): 122.
doi: 10.1186/s12870-021-02879-w pmid: 33648455 |
| [15] | Zhou Y, Xu DX, Jia LD, et al. Genome-wide identification and structural analysis of bZIP transcription factor genes in Brassica napus[J]. Genes, 2017, 8(10): 288. |
| [16] | Jin MJ, Gan SF, Jiao JQ, et al. Genome-wide analysis of the bZIP gene family and the role of AchnABF1 from postharvest kiwifruit(Actinidia chinensis cv. Hongyang)in osmotic and freezing stress adaptations[J]. Plant Sci, 2021, 308: 110927. |
| [17] |
Zou MJ, Guan YC, Ren HB, et al. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance[J]. Plant Mol Biol, 2008, 66(6): 675-683.
doi: 10.1007/s11103-008-9298-4 pmid: 18236009 |
| [18] | Mallappa C, Yadav V, Negi P, et al. A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis[J]. J Biol Chem, 2006, 281(31): 22190-22199. |
| [19] |
Heinekamp T, Kuhlmann M, Lenk A, et al. The tobacco bZIP transcription factor BZI-1 binds to G-box elements in the promoters of phenylpropanoid pathway genes in vitro, but it is not involved in their regulation in vivo[J]. Mol Genet Genomics, 2002, 267(1): 16-26.
doi: 10.1007/s00438-001-0636-3 pmid: 11919711 |
| [20] | Silveira AB, Gauer L, Tomaz JP, et al. The Arabidopsis AtbZIP9 protein fused to the VP16 transcriptional activation domain alters leaf and vascular development[J]. Plant Sci, 2007, 172(6): 1148-1156. |
| [21] | Chuang CF, Running MP, Williams RW, et al. The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana[J]. Genes Dev, 1999, 13(3): 334-344. |
| [22] | Sun XL, Li Y, Cai H, et al. The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses[J]. J Plant Res, 2012, 125(3): 429-438. |
| [23] | Dash M, Yordanov YS, Georgieva T, et al. Poplar PtabZIP1-like enhances lateral root formation and biomass growth under drought stress[J]. Plant J, 2017, 89(4): 692-705. |
| [24] | Kaminaka H, Näke C, Epple P, et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection[J]. EMBO J, 2006, 25(18): 4400-4411. |
| [25] | Jain P, Shah K, Sharma N, et al. A-ZIP53, a dominant negative reveals the molecular mechanism of heterodimerization between bZIP53, bZIP10 and bZIP25 involved in Arabidopsis seed maturation[J]. Sci Rep, 2017, 7(1): 14343. |
| [26] | Llorca CM, Berendzen KW, Malik WA, et al. The elucidation of the interactome of 16 Arabidopsis bZIP factors reveals three independent functional networks[J]. PLoS One, 2015, 10(10): e0139884. |
| [27] | Hossain MA, Cho JI, Han M, et al. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice[J]. J Plant Physiol, 2010, 167(17): 1512-1520. |
| [28] | Bi CX, Yu YH, Dong CH, et al. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat[J]. Plant Biotechnol J, 2021, 19(2): 209-211. |
| [29] |
Wang ZH, Yan LY, Wan LY, et al. Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut[J]. BMC Genomics, 2019, 20(1): 51.
doi: 10.1186/s12864-019-5434-6 pmid: 30651065 |
| [30] | Zhou XJ, Li JT, Wang HL, et al. The chromosome-scale genome assembly, annotation and evolution of Rhododendron henanense subsp. lingbaoense[J]. Mol Ecol Resour, 2022, 22(3): 988-1001. |
| [31] |
Zhou W, Zhang Q, Sun Y, et al. Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses[J]. Int J Biol Macromol, 2020, 145: 341-354.
doi: S0141-8130(19)36966-1 pmid: 31857171 |
| [32] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [33] | Yi SJ, Qian YQ, Han L, et al. Selection of reliable reference genes for gene expression studies in Rhododendron micranthum Turcz[J]. Sci Hortic, 2012, 138: 128-133. |
| [34] |
Schmittgen TD, Livak KJ. Analyzing real-time PCR journal of agricultural biotechnology data by the comparative CT method[J]. Nature Protocols, 2008, 3(6): 1101-1108.
doi: 10.1038/nprot.2008.73 pmid: 18546601 |
| [35] | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana[J]. BMC Plant Biol, 2004, 4: 10. |
| [36] |
Wang SL, Chu ZH, Jia R, et al. SlMYB12 regulates flavonol synthesis in three different cherry tomato varieties[J]. Sci Rep, 2018, 8(1): 1582.
doi: 10.1038/s41598-018-19214-3 pmid: 29371612 |
| [37] | Liu JY, Chen NN, Chen F, et al. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine(Vitis vinifera)[J]. BMC Genomics, 2014, 15: 281. |
| [38] | Hou HY, Kong XJ, Zhou YJ, et al. Genome-wide identification and characterization of bZIP transcription factors in relation to Litchi(Litchi chinensis Sonn.)fruit ripening and postharvest storage[J]. Int J Biol Macromol, 2022, 222(Pt B): 2176-2189. |
| [39] |
刘宝玲, 张莉, 孙岩, 等. 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析[J]. 植物学报, 2016, 51(4): 473-487.
doi: 10.11983/CBB15148 |
| Liu BL, Zhang L, Sun Y, et al. Genome-wide characterization of bZIP transcription factors in foxtail millet and their expression profiles in response to drought and salt stresses[J]. Chin Bull Bot, 2016, 51(4): 473-487. | |
| [40] |
Wei KF, Chen J, Wang YM, et al. Genome-wide analysis of bZIP-encoding genes in maize[J]. DNA Res, 2012, 19(6): 463-476.
doi: 10.1093/dnares/dss026 pmid: 23103471 |
| [41] |
叶方婷, 潘鑫峰, 毛志君, 等. 睡莲转录因子bZIP家族的分子进化以及功能分析[J]. 中国农业科学, 2021, 54(21): 4694-4708.
doi: 10.3864/j.issn.0578-1752.2021.21.018 |
| Ye FT, Pan XF, Mao ZJ, et al. Molecular evolution and function analysis of bZIP family in Nymphaea colorata[J]. Sci Agric Sin, 2021, 54(21): 4694-4708. | |
| [42] | Azeem F, Tahir H, Ijaz U, et al. A genome-wide comparative analysis of bZIP transcription factors in G. arboreum and G. raimondii(Diploid ancestors of present-day cotton)[J]. Physiol Mol Biol Plants, 2020, 26(3): 433-444. |
| [43] |
Zhu M, Yan BW, Hu YJ, et al. Genome-wide identification and phylogenetic analysis of rice FTIP gene family[J]. Genomics, 2020, 112(5): 3803-3814.
doi: S0888-7543(20)30010-0 pmid: 32145381 |
| [44] | Jin ZW, Xu W, Liu AZ. Genomic surveys and expression analysis of bZIP gene family in castor bean(Ricinus communis L.)[J]. Planta, 2014, 239(2): 299-312. |
| [45] | Nuruzzaman M, Manimekalai R, Sharoni AM, et al. Genome-wide analysis of NAC transcription factor family in rice[J]. Gene, 2010, 465(1/2): 30-44. |
| [46] | 郭树娟, 孙月, 郑昊元, 等. 小麦ABF/AREB/ABI5基因家族全基因组鉴定与表达分析[J]. 农业生物技术学报, 2023, 31(4): 667-681. |
| Guo SJ, Sun Y, Zheng HY, et al. Genome-wide identification and expression analysis of ABF/AREB/ABI5 gene family in wheat(Triticum aestivum)[J]. J Agric Biotechnol, 2023, 31(4): 667-681. | |
| [47] | Murmu J, Bush MJ, DeLong C, et al. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development[J]. Plant Physiol, 2010, 154(3): 1492-1504. |
| [48] |
Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants[J]. New Phytol, 2009, 183(3): 557-564.
doi: 10.1111/j.1469-8137.2009.02923.x pmid: 19555435 |
| [49] |
Zhang M, Liu YH, Shi H, et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family[J]. BMC Genomics, 2018, 19(1): 159.
doi: 10.1186/s12864-018-4511-6 pmid: 29471787 |
| [50] | 孙耀国, 蔡天润, 姬行舟, 等. 西洋梨全基因组bZIP基因家族生物信息学分析[J]. 林业与生态科学, 2021, 36(1): 24-34. |
| Sun YG, Cai TR, Ji XZ, et al. Genome-wide bioinformatics analysis of bZIP gene family in Pyrus communis[J]. For Ecol Sci, 2021, 36(1): 24-34. | |
| [51] |
Chi YH, Cheng YS, Vanitha J, et al. Expansion mechanisms and functional divergence of the glutathione s-transferase family in sorghum and other higher plants[J]. DNA Res, 2011, 18(1): 1-16.
doi: 10.1093/dnares/dsq031 pmid: 21169340 |
| [52] |
Hörberg J, Reymer A. Specifically bound BZIP transcription factors modulate DNA supercoiling transitions[J]. Sci Rep, 2020, 10(1): 18795.
doi: 10.1038/s41598-020-75711-4 pmid: 33139763 |
| [53] | Zhao P, Ye MH, Wang RQ, et al. Systematic identification and functional analysis of potato(Solanum tuberosum L.)bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance[J]. Int J Biol Macromol, 2020, 161: 155-167. |
| [54] | 杨芳, 杨仕梅, 罗雪, 等. 百脉根Hsp70s基因家族的生物信息学分析[J]. 山地农业生物学报, 2020, 39(5): 1-8. |
| Yang F, Yang SM, Luo X, et al. Bioinformatics analysis of Hsp70s gene family in Lotus japonicus[J]. J Mt Agric Biol, 2020, 39(5): 1-8. | |
| [55] | Yang ZM, Sun J, Chen Y, et al. Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida[J]. BMC Genet, 2019, 20(1): 41. |
| [56] | Xu DB, Chen M, Ma YN, et al. A G-protein β subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis[J]. PLoS One, 2015, 10(1): e0116385. |
| [57] | Liu DF, Shi SP, Hao ZJ, et al. OsbZIP81, a homologue of Arabidopsis VIP1, may positively regulate JA levels by directly targeting the genes in JA signaling and metabolism pathway in rice[J]. Int J Mol Sci, 2019, 20(9): 2360. |
| [58] |
Gangappa SN, Botto JF. The multifaceted roles of HY5 in plant growth and development[J]. Mol Plant, 2016, 9(10): 1353-1365.
doi: S1674-2052(16)30134-4 pmid: 27435853 |
| [59] |
Yang Y, Liang T, Zhang LB, et al. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis[J]. Nat Plants, 2018, 4(2): 98-107.
doi: 10.1038/s41477-017-0099-0 pmid: 29379156 |
| [60] | van Gelderen K, Kang C, Paalman R, et al. Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor[J]. Plant Cell, 2018, 30(1): 101-116. |
| [61] | Li QF, He JX. BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated Cotyledon opening in Arabidopsis in darkness[J]. Mol Plant, 2016, 9(1): 113-125. |
| [62] | Yu YW, Wang J, Zhang ZJ, et al. Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light[J]. PLoS Genet, 2013, 9(12): e1004025. |
| [63] |
Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance[J]. Plant Cell Physiol, 2011, 52(9): 1569-1582.
doi: 10.1093/pcp/pcr106 pmid: 21828105 |
| [64] |
Seo PJ. Recent advances in plant membrane-bound transcription factor research: emphasis on intracellular movement[J]. J Integr Plant Biol, 2014, 56(4): 334-342.
doi: 10.1111/jipb.12139 |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||