








生物技术通报 ›› 2021, Vol. 37 ›› Issue (8): 111-120.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1591
刘媛媛( ), 杨冬杰, 左东云, 程海亮, 张友平, 吕丽敏, 王巧连, 宋国立(
), 杨冬杰, 左东云, 程海亮, 张友平, 吕丽敏, 王巧连, 宋国立( )
)
收稿日期:2020-12-31
出版日期:2021-08-26
发布日期:2021-09-10
作者简介:刘媛媛,女,硕士,研究方向:作物遗传育种;E-mail: 基金资助:
LIU Yuan-yuan( ), YANG Dong-jie, ZUO Dong-yun, CHENG Hai-liang, ZHANG You-ping, LV Li-min, WANG Qiao-lian, SONG Guo-li(
), YANG Dong-jie, ZUO Dong-yun, CHENG Hai-liang, ZHANG You-ping, LV Li-min, WANG Qiao-lian, SONG Guo-li( )
)
Received:2020-12-31
Published:2021-08-26
Online:2021-09-10
摘要:
旨在探究丝氨酸苏氨酸蛋白激酶(serine/ threonine protein kinase,STPK)基因在棉纤维发育中的功能,解析棉纤维细胞的分化和发育机理。从陆地棉遗传标准系TM-1中克隆基因GhD6PKL2,并对其序列及结构特征进行生物信息学分析、表达量分析及过表达拟南芥的表型观察。GhD6PKL2含有典型的STPK保守序列位点。预测编码的蛋白相对分子质量为49.74 kD,等电点为6.17,含有多个丝氨酸苏氨酸磷酸化位点。表达模式结果表明,GhD6PKL2在与棉花纤维伸长阶段相吻合的、开花后20 d显著高表达。软件预测及在烟草叶片中的荧光蛋白定位结果均显示,GhD6PKL2编码的蛋白质定位在细胞膜上。诱饵自激活试验验证GhD6PKL2没有自激活活性及毒性。过表达拟南芥,能使转基因拟南芥表现出表皮毛数量增多,同时主根变短、侧根数目增多的表型。表明该丝氨酸苏氨酸蛋白激酶基因在拟南芥表皮毛及主侧根发育方面发挥一定作用,可能为一个棉纤维伸长发育阶段的潜在调控基因。
刘媛媛, 杨冬杰, 左东云, 程海亮, 张友平, 吕丽敏, 王巧连, 宋国立. 棉花GhD6PKL2的克隆及功能验证[J]. 生物技术通报, 2021, 37(8): 111-120.
LIU Yuan-yuan, YANG Dong-jie, ZUO Dong-yun, CHENG Hai-liang, ZHANG You-ping, LV Li-min, WANG Qiao-lian, SONG Guo-li. Cloning and Functional Verification of GhD6PKL2 from Gossypium hirsutum[J]. Biotechnology Bulletin, 2021, 37(8): 111-120.
| 引物用途 Function of the primer | 引物名称 Name of the primer | 引物序列 Sequence of the primer(5'-3') |
|---|---|---|
| pRI101载体通用引物 The primers in pRI101 vector | 35ScexuPF | CCTTCGCAAGACCCTTCCTC |
| pRI101- AN- cexuRV | CAGGAAACAGCTATGAC | |
| cDNA扩增& 过表达载体构建引物 The primers for cDNA amplification and over-expression vector’s construction | D6PKPFCZ(Sal I) | TTGATACATATGCCCGTCGACATGGAGCCGTTTCTCGACGACT |
| D6PKPR1CZ(EcoR I) | GAATTCCTAATGATGATGATGATGATGATAATACTCTACTGGGGTCTCT | |
| D6PKPR2CZ(EcoR I) | AGAGTTGTTGATTCAGAATTCCTAATGATGATGATGATGATG | |
| 定量引物 The primers for quantitative real-time PCR | A07GD6PKPF1 | TGAGTTCCCTAAAGAACCCATTGT |
| A07GD6PKPR1 | GGTGGTGCTTGATGGCTGAT | |
| cDNA检测& 内参引物 The primers for cDNA quality’s verification and reference | AtactinePF | TGCTATTCTGCGTTTGGACCTTG |
| AtactinePR | ATCCCTTACGATTTCACGCTCTG | |
| Histone3PF1 | CCGTAAATCTGCCCCAACCA | |
| Histone3PR1 | GACCCACAAGGTATGCCTCTGC | |
| GFP& Linker片段扩增引物 The primers for GFP and Linker fragments amplification | GFPlinkPF1 | TGGCTCTGGCGGTGGCGGATCGATGGGTAAAGGAGAAGAACTTT |
| GFPPR1(EcoR I) | AGAGTTGTTGATTCAGAATTCTCATTTGTATAGTTCATCCATG | |
| LinkPF2(BamH I) | GGATCCGGTGGAGGCGGTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCG | |
| 亚细胞定位中基因扩增引物 The primers for genes in subcellular location | AD6PKPF1(Sal I) | TTGATACATATGCCCGTCGACATGGAGCCGTTTCTCGACGACT |
| AD6PKPR1(BamH I) | CTGAACCGCCTCCACCGGATCCATAATACTCTACTGGGGTCTCT | |
| 诱饵载体引物 The primers for bait vector construction | A0D6PKPF1GB(Sal) | ATGCGGCCGCTGCAGGTCGACATGGAGCCGTTTCTCGACGACT |
| A0D6PKPR1GB(EcoR) | ATGGCCATGGAGGCCGAATTCATAATACTCTACTGGGGTCTCT |
表1 本研究所用到的引物列表
Table1 Primers used in this study
| 引物用途 Function of the primer | 引物名称 Name of the primer | 引物序列 Sequence of the primer(5'-3') |
|---|---|---|
| pRI101载体通用引物 The primers in pRI101 vector | 35ScexuPF | CCTTCGCAAGACCCTTCCTC |
| pRI101- AN- cexuRV | CAGGAAACAGCTATGAC | |
| cDNA扩增& 过表达载体构建引物 The primers for cDNA amplification and over-expression vector’s construction | D6PKPFCZ(Sal I) | TTGATACATATGCCCGTCGACATGGAGCCGTTTCTCGACGACT |
| D6PKPR1CZ(EcoR I) | GAATTCCTAATGATGATGATGATGATGATAATACTCTACTGGGGTCTCT | |
| D6PKPR2CZ(EcoR I) | AGAGTTGTTGATTCAGAATTCCTAATGATGATGATGATGATG | |
| 定量引物 The primers for quantitative real-time PCR | A07GD6PKPF1 | TGAGTTCCCTAAAGAACCCATTGT |
| A07GD6PKPR1 | GGTGGTGCTTGATGGCTGAT | |
| cDNA检测& 内参引物 The primers for cDNA quality’s verification and reference | AtactinePF | TGCTATTCTGCGTTTGGACCTTG |
| AtactinePR | ATCCCTTACGATTTCACGCTCTG | |
| Histone3PF1 | CCGTAAATCTGCCCCAACCA | |
| Histone3PR1 | GACCCACAAGGTATGCCTCTGC | |
| GFP& Linker片段扩增引物 The primers for GFP and Linker fragments amplification | GFPlinkPF1 | TGGCTCTGGCGGTGGCGGATCGATGGGTAAAGGAGAAGAACTTT |
| GFPPR1(EcoR I) | AGAGTTGTTGATTCAGAATTCTCATTTGTATAGTTCATCCATG | |
| LinkPF2(BamH I) | GGATCCGGTGGAGGCGGTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCG | |
| 亚细胞定位中基因扩增引物 The primers for genes in subcellular location | AD6PKPF1(Sal I) | TTGATACATATGCCCGTCGACATGGAGCCGTTTCTCGACGACT |
| AD6PKPR1(BamH I) | CTGAACCGCCTCCACCGGATCCATAATACTCTACTGGGGTCTCT | |
| 诱饵载体引物 The primers for bait vector construction | A0D6PKPF1GB(Sal) | ATGCGGCCGCTGCAGGTCGACATGGAGCCGTTTCTCGACGACT |
| A0D6PKPR1GB(EcoR) | ATGGCCATGGAGGCCGAATTCATAATACTCTACTGGGGTCTCT |

图1 GhD6PKL2序列分析及蛋白比对 A:GhD6PKL2结构示意图;B:GhD6PKL2与其他几个物种(AtD6PKL2-1:拟南芥;NtD6PKL2-11:烟草;OsD6PKL2-1:水稻;ZmHPK-1:玉米)氨基酸序列的多重比对。图中标注罗马数字Ⅰ-Ⅺ中处为STPK家族中11个典型的活性位点
Fig.1 Analysis of the GhD6PKL2 sequence and protein sequence alignment A: The structure of the GhD6PKL2. B: Multiple alignment of amino acid sequences between GhD6PKL2 and other’s species(AtD6PKL2- 1: Arabidopsis; NtD6PKL2- 11: Nicotiana tabacum; OsD6PKL2-1: Oryza sativa; ZmHPK-1: Zea mays). The number Ⅰ to Ⅺ indicate 11 typical active sites of STPK family
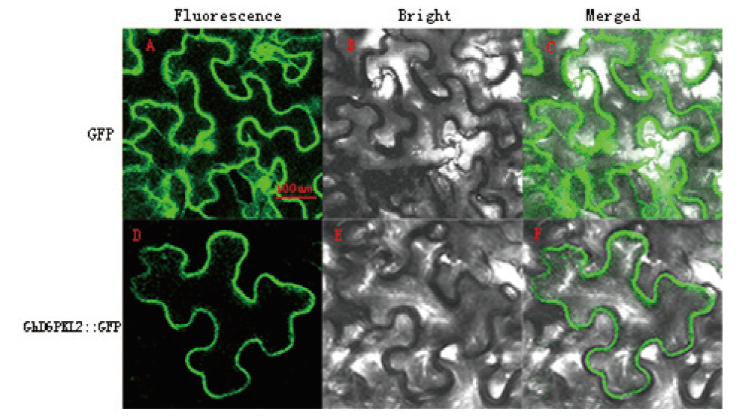
图4 GhD6PKL2的亚细胞定位 A-C:对照;D-F:GhD6PKL2-GFP融合蛋白;bar=100 µm
Fig.4 Subcellular localization of GhD6PKL2 A- C:GFP only; D- F: the fusion protein: GhD6PKL2- GFP; bar: 100 m
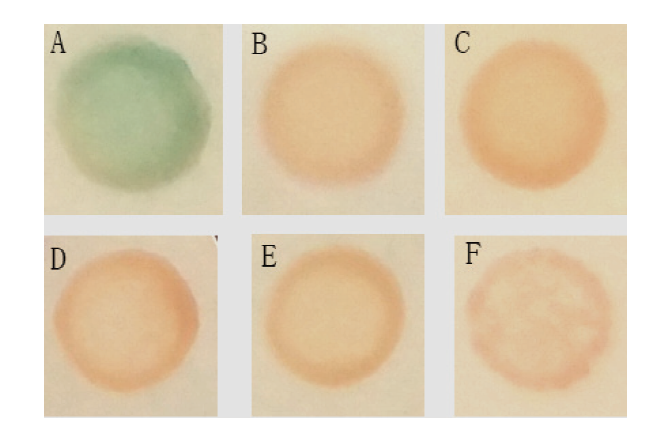
图5 GhD6PKL2诱饵载体自激活的检测 A:阳性对照pGBKT7-53和pGADT7-T在SD-trp-leu/X-α-gal培养基上的生长情况;B:阴性对照pGBKT7-Lam和pGADT7-T在SD-trp-leu/X-α-gal培养基上的生长情况;C:空白对照PGADT7-T在SD-trp培养基上的生长情况;D:pGBKT7-GhD6PKL2在SD-trp培养基上的生长情况;E:pGBKT7-GhD6PKL2在SD-trp/X-α-gal培养基上的生长情况;F:pGBKT7-GhD6PKL2在SD-trp/X-α-gal/AbA培养基上的生长情况
Fig.5 Detection of transcriptional activity of GhD6PKL2 A: Positive control: pGBKT7-53 and pGADT7-T, growth on SD-trp-leu/X-α-gal;B: nagetive control: pGBKT7-Lam and pGADT7-T growth on SD-trp-leu/X-α-gal; C: blank control: PGADT7-T growth on SD-trp; D: pGBKT7-GhD6PKL2 growth on SD-trp; E: pGBKT7-GhD6PKL2 growth on SD-trp/X-α-gal; F: pGBKT7-GhD6PKL2 growth on SD-trp/X-α-gal/AbA

图7 转GhD6PKL2拟南芥叶片表皮毛的表型 A:转GhD6PKL2拟南芥和WT表皮毛观察;B:转GhD6PKL2拟南芥和WT表皮毛数量平均值。***表示在P=0.001水平差异显著。下同
Fig.7 Phynotype of Arabidopsis trichome between GhD6PKL2 trans-genic plants and wide type A: Arabidopsis trichome observation between transgenic plants and wide type;B: average arabidopsis trichome in transgenic plants and wide type. ***indicate that there is a significant difference at P= 0.001. The same below
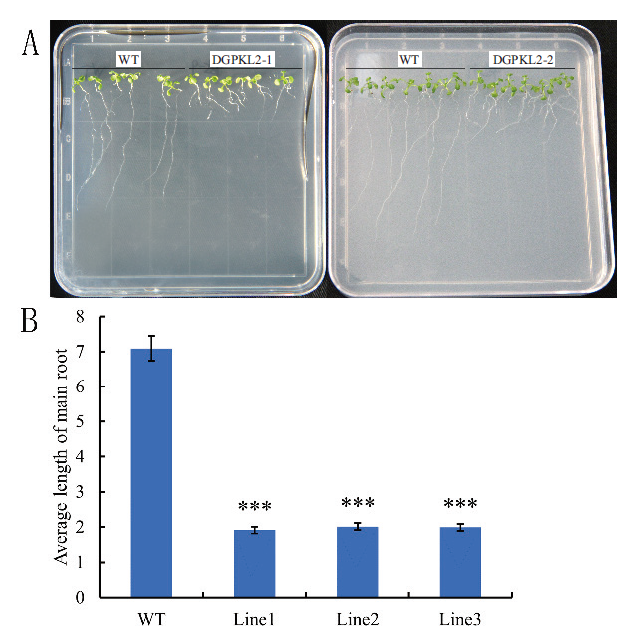
图8 转GhD6PKL2拟南芥根的表型 A:转GhD6PKL2拟南芥和WT根生长情况;B:转GhD6PKL2拟南芥和WT主根长度
Fig.8 Phenotype of Arabidopsis main root between GhD6PKL2 trans-genic plants and wide type A: Arabidopsis root growths of GhD6PKL2 transgenic plants and WT type; B: Main root length in GhD6PKL2 transgenic plants and WT type.
| [1] |
Pitzschke A, Schikora A, Hirt H. MAPK cascade signalling networks in plant defence[J]. Curr Opin Plant Biol, 2009, 12(4):421-426.
doi: 10.1016/j.pbi.2009.06.008 pmid: 19608449 |
| [2] |
Goldberg J, Huang HB, Kwon YG, et al. Three- dimensional structure of the catalytic subunit of protein serine/threonine phosphatase- 1[J]. Nature, 1995, 376(6543):745-753.
doi: 10.1038/376745a0 URL |
| [3] |
Hanks SK, Quinn AM, Hunter T. The protein-kinase family:conserved features and deduced phylogeny of the catalytic domains[J]. Science, 1988, 241(4861):42-52.
pmid: 3291115 |
| [4] | Rademacher EH, Offringa R. Evolutionary adaptations of plant AGC kinases:from light signaling to cell polarity regulation[J]. Frontiers in Plant Science, 2012, 3(250). |
| [5] |
Huse M, Kuriyan J. The conformational plasticity of protein kinases[J]. Cell, 2002, 109(3):275-282.
doi: 10.1016/S0092-8674(02)00741-9 URL |
| [6] |
Hanks SK, Hunter T. The eukaryotic protein kinase superfamily:kinase(catalytic)domain structure and classification[J]. The FASEB Journal, 1995, 9(8):576-596.
doi: 10.1096/fsb2.v9.8 URL |
| [7] |
Bogre L, Okresz L, Henriques R, et al. Growth signalling pathways in Arabidopsis and the AGC protein kinases[J]. Trends in Plant Science, 2003, 8(9):424-431.
doi: 10.1016/S1360-1385(03)00188-2 URL |
| [8] | Zourelidou M, Muller I, Willige BC, et al. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana[J]. Develop, 2009, 136(4):627-636. |
| [9] |
Galvan-Ampudia CS, Offringa R. Plant evolution:AGC kinases tell the auxin tale[J]. Trends Plant Sci, 2007, 12(12):541-547.
doi: 10.1016/j.tplants.2007.10.004 URL |
| [10] |
Sakai T, Kagawa T, Kasahara M, et al. Arabidopsis nph1 and npl1:blue light receptors that mediate both phototropism and chloroplast relocation[J]. PNAS, 2001, 98(12):6969-6974.
pmid: 11371609 |
| [11] | Briggs WR, Huala E. Blue- light photoreceptors in higher plants[J]. Annu Rev Cell Devl Biol, 1999, 15:33-62. |
| [12] |
Santner AA, Watson JC. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis[J]. The Plant Journal, 2010, 45(5):752-764.
doi: 10.1111/tpj.2006.45.issue-5 URL |
| [13] |
Haga K, Hayashi KI, Sakai T. PINOID AGC kinases are necessary for phytochrome- mediated enhancement of hypocotyl phototropism in Arabidopsis[J]. Plant Physiol, 2014, 166(3):1535-1545.
doi: 10.1104/pp.114.244434 pmid: 25281709 |
| [14] |
Zhang Y, He JM, Mccormick S. Two Arabidopsis AGC kinases are critical for the polarized growth of pollen tubes[J]. The Plant Journal, 2010, 58(3):474-484.
doi: 10.1111/tpj.2009.58.issue-3 URL |
| [15] |
Enugutti B, Kirchhelle C, Oelschner M, et al. Regulation of planar growth by the Arabidopsis AGC protein kinase UNICORN[J]. Proc Natl Acad Sci USA, 2012, 109(37):15060-15065.
doi: 10.1073/pnas.1205089109 URL |
| [16] |
Haga K, Frank L, Kimura T, et al. Roles of AGC VIII kinases in the hypocotyl phototropism of Arabidopsis seedlings[J]. Plant Cell Physiology, 2018, 59(5):1060-1071.
doi: 10.1093/pcp/pcy048 URL |
| [17] |
Devarenne TP, Ekengren SK, Pedley KF, et al. Adi3 is a PDK1- interacting AGC kinase that negatively regulates plant cell death[J]. The EMBO Journal, 2006, 25(1):255-265.
doi: 10.1038/sj.emboj.7600910 URL |
| [18] |
Hammond RW, Zhao Y. Modification of tobacco plant development by sense and antisense expression of the tomato viroid- induced AGC VIIIa protein kinase PKV suggests involvement in gibberellin signaling[J]. BMC Plant Biology, 2009, 9:108.
doi: 10.1186/1471-2229-9-108 URL |
| [19] |
Zhang YY, et al. AGC protein kinase AGC1- 4 mediates seed size in Arabidopsis[J]. Plant Cell Rep, 2020, 39(6):825-837.
doi: 10.1007/s00299-020-02533-z URL |
| [20] |
Wan Q, Guan XY, Yang NN, et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development[J]. New Phytologist, 2016, 210(4):1298-1310.
doi: 10.1111/nph.2016.210.issue-4 URL |
| [21] |
Serna L, Martin C. Trichomes:different regulatory networks lead to convergent structures[J]. Trends Plant Sci, 2006, 11(6):274-280.
doi: 10.1016/j.tplants.2006.04.008 URL |
| [22] |
Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development[J]. Annals of Botany, 2008, 100(7):1391-1401.
doi: 10.1093/aob/mcm232 URL |
| [23] |
Guan XY, Pang MX, Nah G, et al. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development[J]. Nat Commun, 2014, 5:3050.
doi: 10.1038/ncomms4050 URL |
| [24] |
Wang S, Wang JW, Yu N, et al. Control of plant trichome development by a cotton fiber MYB gene[J]. The Plant Cell, 2004, 16(9):2323-2334.
doi: 10.1105/tpc.104.024844 URL |
| [25] |
Guan XY, Lee JJ, Pang MX, et al. Activation of Arabidopsis seed hair development by cotton fiber- related genes[J]. PLoS One, 2011 6(7):e21301.
doi: 10.1371/journal.pone.0021301 URL |
| [26] |
Zourelidou M, Absmanner B, Weller B, et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID[J]. eLife, 2014, 3:e02860.
doi: 10.7554/eLife.02860 URL |
| [27] |
Willige BC, Ahlers S, Zourelidou M, et al. D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis[J]. The Plant Cell, 2013, 25(5):1674-1688.
doi: 10.1105/tpc.113.111484 pmid: 23709629 |
| [28] | Willige BC, Joanne C. A current perspective on the role of AGCVIII kinases in PIN- mediated apical hook development[J]. Frontiers in Plant science, 2015, 6(767):767-773. |
| [29] |
Stanislas T, Huser A, Barbosa IC, et al. Arabidopsis D6PK is a lipid domain- dependent mediator of root epidermal planar polarity[J]. Nature Plants, 2015, 1(11):15162.
doi: 10.1038/nplants.2015.162 URL |
| [30] |
Lee BH, Weber ZT, et al. Arabidopsis protein kinase D6PKL3 is involved in the formation of distinct plasma membrane aperture domains on the pollen surface[J]. Plant Cell, 2018, 30(9):2038-2056.
doi: 10.1105/tpc.18.00442 URL |
| [31] |
Daisuke M, Takashi N, Sato KI, et al. Activation of AtMEK1, an Arabidopsis mitogen- activated protein kinase kinase, in vitro and in vivo:analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings[J]. The Plant Journal, 2002, 29(5):637-647.
doi: 10.1046/j.0960-7412.2001.01246.x URL |
| [32] |
Simon ML, Platre MP, Marques-Bueno MM, et al. A PtdIns(4)P- driven electrostatic field controls cell membrane identity and signalling in plants[J]. Nature Plants, 2016, 2:16089.
doi: 10.1038/nplants.2016.89 URL |
| [33] |
Ek-Ramos MJ, Julian A, Nelson-Dittrich AC, et al. The tomato cell death suppressor Adi3 is restricted to the endosomal system in response to the Pseudomonas syringae effector protein AvrPto[J]. PLoS One, 2014, 9(10):e110807.
doi: 10.1371/journal.pone.0110807 URL |
| [34] |
Xiao Y, Offringa R. PDK1 regulates auxin transport and Arabidopsis vascular development through AGC1 kinase PAX[J]. Nature Plants, 2020, 6(5):1-12.
doi: 10.1038/s41477-019-0586-6 URL |
| [35] |
Zeng JY, Zhang M, Hou L, et al. Cytokinin inhibits cotton fiber initiation by disrupting PIN3a- mediated asymmetric accumulation of auxin in the ovule epidermis[J]. Journal of Experimental Botany, 2019, 70(12):3139-3151.
doi: 10.1093/jxb/erz162 URL |
| [36] | Benjamins R, Quint A, et al. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport[J]. Develop, 2001, 128(20):4057-4067. |
| [37] | Weller B, Zourelidou M, Frank L, et al. Dynamic PIN- FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux- dependent growth[J]. Proc Natl Acad Sci. USA, 2017, 114(5):201614380. |
| [38] |
Tan ST, Zhang XX, Kong W, et al. A lipid code-dependent phosphoswitch directs PIN- mediated auxin efflux in Arabidopsis development[J]. Nature Plants, 2020, 6(5):556-569.
doi: 10.1038/s41477-020-0648-9 URL |
| [39] |
Zhu QK, Shao YM, Ge ST, et al. A MAPK cascade downstream of IDA-HAE/ HSL2 ligand- receptor pair in lateral root emergence[J]. Nature Plants, 2019, 5:414-423.
doi: 10.1038/s41477-019-0396-x URL |
| [40] |
Marhava P, Bassukas AE, Zourelidou M, et al. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation[J]. Nature, 2018, 558(7709):297-300.
doi: 10.1038/s41586-018-0186-z URL |
| [41] |
Oyama T, Shimura Y, Okada K. The IRE gene encodes a protein kinase homologue and modulates root hair growth in Arabidopsis[J]. The Plant Journal, 2002, 30(3):289-299.
doi: 10.1046/j.1365-313X.2002.01290.x URL |
| [42] |
Anthony RG, Khan S, Costa J, et al. The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1[J]. Journal of Biological Chemistry, 2006, 281(49):37536-37546.
doi: 10.1074/jbc.M607341200 URL |
| [1] | 王一帆, 候林慧, 常永春, 杨亚杰, 陈天, 赵祝跃, 荣二花, 吴玉香. 陆地棉与拟似棉异源六倍体的合成与性状鉴定[J]. 生物技术通报, 2023, 39(5): 168-176. |
| [2] | 刘萌萌, 韩立军, 刘宝玲, 薛金爱, 李润植. 陆地棉GhSDP1及其启动子的克隆与表达分析[J]. 生物技术通报, 2022, 38(2): 34-43. |
| [3] | 马麒, 李吉莲, 徐守振, 陈红, 刘文豪, 宁新柱, 林海. 陆地棉果枝夹角性状的主基因+多基因混合遗传模型分析[J]. 生物技术通报, 2022, 38(10): 148-158. |
| [4] | 杨笑敏, 王俊娟, 王德龙, 陆许可, 陈修贵, 郭丽雪, 王帅, 陈超, 王晓歌, 韩明格, 叶武威. 棉花GhDMT3的功能验证及生物信息学分析[J]. 生物技术通报, 2019, 35(1): 11-16. |
| [5] | 刘妍, 孟志刚, 孙国清, 王远, 周焘, 郭三堆, 张锐. 陆地棉GhPYR1基因的克隆和功能分析[J]. 生物技术通报, 2016, 32(2): 90-99. |
| [6] | 徐珍珍, 倪万潮, 张香桂, 郭琪, 徐鹏, 沈新莲. 棉花YABBY基因家族的全基因组分析[J]. 生物技术通报, 2015, 31(11): 146-152. |
| [7] | 胡根海;王清连;郭敏敏;张金宝;胡存科;. 陆地棉子叶节高效再生体系的建立[J]. , 2009, 0(06): 109-111. |
| [8] | 孙志栋;王学德;倪西源;. 棉花2个多标记基因系及其杂交后代AFLP分析[J]. , 2006, 0(S1): 293-295. |
| [9] | . 文摘[J]. , 2001, 0(05): 53-55. |
| [10] | 孙雷心. 抗生素能否清除植物组培物中的细菌[J]. , 1992, 0(08): 6-7. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||