








生物技术通报 ›› 2022, Vol. 38 ›› Issue (5): 269-278.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0892
收稿日期:2021-07-12
出版日期:2022-05-26
发布日期:2022-06-10
作者简介:朱秋雨,女,硕士研究生,研究方向:食品与发酵工程;E-mail: 基金资助:Received:2021-07-12
Published:2022-05-26
Online:2022-06-10
摘要:
将枯草芽孢杆菌L-天冬氨酸-α-脱羧酶基因进行了克隆和异源表达,并通过定点突变构建了2个突变体。针对该酶活力检测时存在的检测通量低、周期长和成本高等缺点,旨在建立一种简单高效的酶活力高通量检测方法。采用氯酚红(CPR)指示剂和4-吗啉乙磺酸(MES)缓冲液体系,并对检测条件进行优化,提高检测的准确性和灵敏性,建立了基于比色法的微孔板高通量检测方法,然后以L-天冬氨酸-α-脱羧酶及其突变体作为模型酶,对高通量检测方法进行了验证。优化后的酶活检测条件为MES缓冲液2 mmol/L,CPR指示剂75 μmol/L,L-天冬氨酸75 mmol/L,pH 6.5,温度37℃,反应时间10 min,检测波长为567 nm。采用3种模型酶对微孔板高通量检测方法进行了验证,结果显示该方法与HPLC法测得的结果一致。高通量检测方法具有操作简便易行、灵敏度高等优点,能够用于L-天冬氨酸-α-脱羧酶的快速检测。该方法的建立将为L-天冬氨酸-α-脱羧酶进行定向进化及突变体的高通量筛选奠定基础。
朱秋雨, 段绪果. L-天冬氨酸-α-脱羧酶的重组表达、定点突变及高通量检测方法的建立[J]. 生物技术通报, 2022, 38(5): 269-278.
ZHU Qiu-yu, DUAN Xu-guo. Recombinant Expression and Site-directed Mutagenesis of L-aspartate-α-decarboxylase,and the Establishment of High-throughput Assay Method[J]. Biotechnology Bulletin, 2022, 38(5): 269-278.
| 引物Primer | 引物序列Primer sequence(5'-3') |
|---|---|
| BspanD-F | GAAGGAGATATACATATGTATCGAACAATGATGAGCGG |
| BspanD-R | GCTTGTCGACGGAGCTCCTACAAAATTGTACGGGCTGGT |
| I88M-F | GGTCATTATTATGTCCCACAAAATGATGTC |
| I88M-R | GACATCATTTTGTGGGACATAATAATGACC |
| I46V-F | GAAAAAGTACAAGTTGTGAATAATAATAATGGAG |
| I46V-R | CTCCATTATTATTATTCACAACTTGTACTTTTTC |
| K104S-F | CCATGAGCCGAGTGTGGCTGTTCT |
| K104S-R | AGAACAGCCACACTCGGCTCATGG |
| I126*-F | GAACCAGCCCGTACATAATTGTAGGAGC |
| I126*-R | GCTCCTACAATTATGTACGGGCTGGTTC |
表1 本研究中使用的引物
Table 1 Primers used in this study
| 引物Primer | 引物序列Primer sequence(5'-3') |
|---|---|
| BspanD-F | GAAGGAGATATACATATGTATCGAACAATGATGAGCGG |
| BspanD-R | GCTTGTCGACGGAGCTCCTACAAAATTGTACGGGCTGGT |
| I88M-F | GGTCATTATTATGTCCCACAAAATGATGTC |
| I88M-R | GACATCATTTTGTGGGACATAATAATGACC |
| I46V-F | GAAAAAGTACAAGTTGTGAATAATAATAATGGAG |
| I46V-R | CTCCATTATTATTATTCACAACTTGTACTTTTTC |
| K104S-F | CCATGAGCCGAGTGTGGCTGTTCT |
| K104S-R | AGAACAGCCACACTCGGCTCATGG |
| I126*-F | GAACCAGCCCGTACATAATTGTAGGAGC |
| I126*-R | GCTCCTACAATTATGTACGGGCTGGTTC |
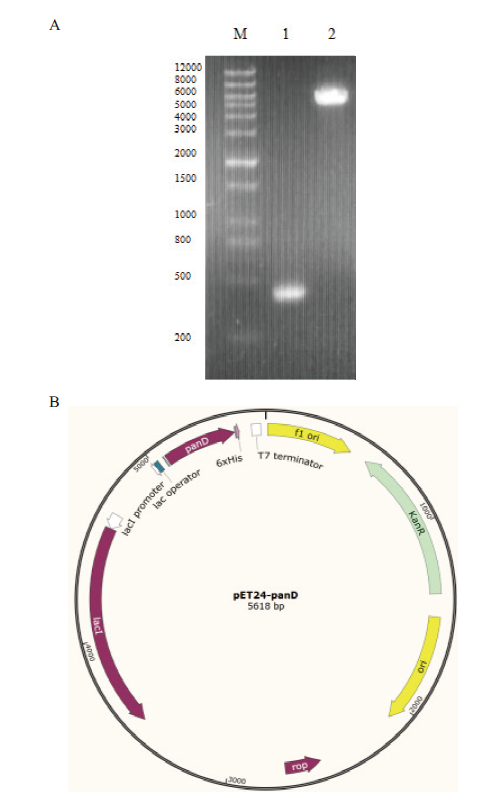
图1 琼脂糖凝胶电泳图及表达载体结构示意图 A:panDBs 基因PCR产物和质粒酶切产物(M:标准DNA分子量;1:panDBs PCR产物;2:质粒pET24a(+)双酶切产物);B:重组表达载体pET24a(+)-panDBs示意图
Fig. 1 Agarose gel electropherogram and structural sch-ematic diagram of expression vector A:PCR product of panDBs and plasmid enzyme digestion product(M:DNA marker;1:PCR product of panDBs;2:double restriction enzyme digestion product of plasmid pET24a(+)). B:Structural schematic diagram of recombinant expression vector pET24a(+)-panDBs
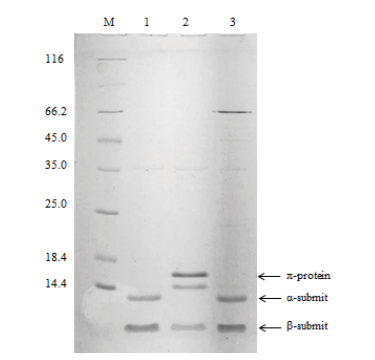
图2 重组酶panDBs及突变体蛋白电泳图 M:标准蛋白分子量;1:panDBs上清;2:突变体panDBs-1上清;3:突变体panDBs-2上清
Fig. 2 Electropherogram of recombinant protein panDBs and its mutant proteins M:Protein marker. 1:Supernatant of panDBs. 2:Supernatant of mutant panDBs-1. 3:Supernatant of mutant panDBs-2
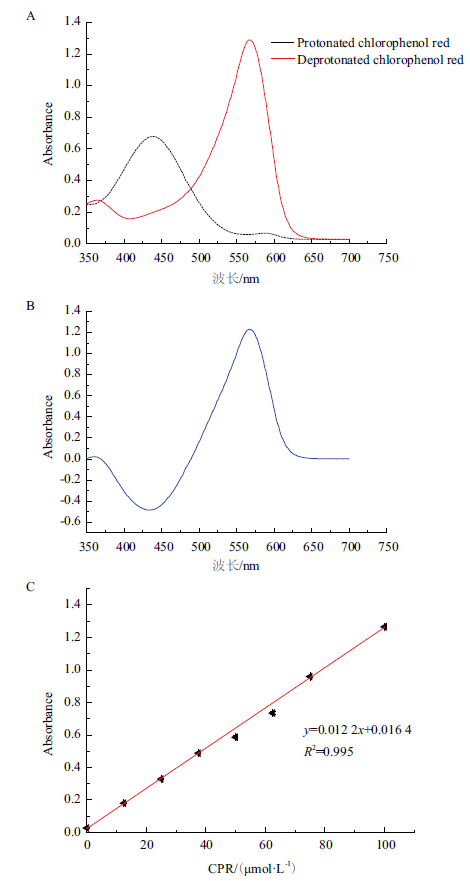
图4 基于比色法的PanD高通量筛选方法的建立 A:质子化和去质子化的CPR吸收光谱;B:质子化和去质子化的吸收光谱差异;C:567 nm处的吸光度与氯酚红(CPR)浓度的关系
Fig. 4 Development of a colorimetric high-throughput scr-eening method for PanD assay A:Absorption spectra of protonated and deprotonated forms of CPR. B:Difference in absorption spectra between the two forms of CPR. C:Absorbance at 567 nm as a function of the concentration of CPR
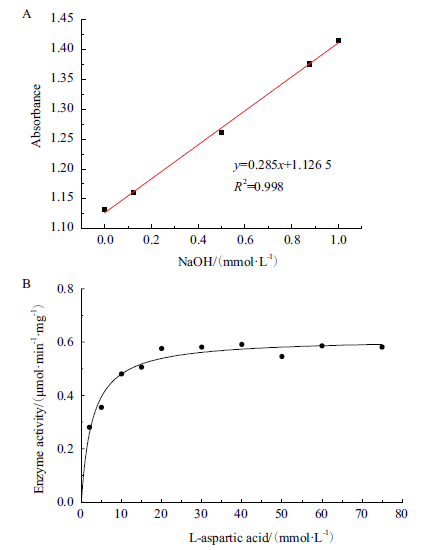
图7 PanD活性的测定 A:微孔板中CPR-MES体系校准曲线;B:酶反应速率曲线
Fig. 7 Determination of PanD activity A:Validation curve of CPR-MES system in a microplate. B:Enzymatic reaction rate curve
| Mutation sites | Vmax/ (μmol·min-1·mg-1) | Km/ (mmol·L-1) | |
|---|---|---|---|
| panDBs | - | 0.61±0.06 | 2.78±0.09 |
| panDBs-1 | I46V/I88M/K104S/I126* | 0.68±0.03 | 2.98±0.16 |
| panDBs-2 | I88M | 1.11±0.11 | 1.49±0.05 |
表2 重组酶及其突变体的部分动力学参数
Table 2 Partial kinetic parameters of recombinant enzyme and its mutants
| Mutation sites | Vmax/ (μmol·min-1·mg-1) | Km/ (mmol·L-1) | |
|---|---|---|---|
| panDBs | - | 0.61±0.06 | 2.78±0.09 |
| panDBs-1 | I46V/I88M/K104S/I126* | 0.68±0.03 | 2.98±0.16 |
| panDBs-2 | I88M | 1.11±0.11 | 1.49±0.05 |
| [1] |
Williamson JM, Brown GM. Purification and properties of L-Aspartate-alpha-decarboxylase, an enzyme that catalyzes the formation of beta-alanine in Escherichia coli[J]. J Biol Chem, 1979, 254(16):8074-8082.
pmid: 381298 |
| [2] | 范海洋. 重组大肠杆菌L-天冬氨酸-α-脱羧酶的制备及应用研究[D]. 上海: 华东理工大学, 2013. |
| Fan HY. Preparation and application of recombinant Escherichia coli L-aspartate-a-decdarboxylase[D]. Shanghai: East China University of Science and Technology, 2013. | |
| [3] | 赵连真, 张梁, 石贵阳. 谷氨酸棒杆菌L-天冬氨酸α-脱羧酶在大肠杆菌中的表达及酶转化生产β-丙氨酸[J]. 微生物学通报, 2013, 40(12):2161-2170. |
| Zhao LZ, Zhang L, Shi GY. Expression of L-aspartate α-decarboxylase from Corynebacterium glutamicum in Escherichia coli and its application in enzymatic synthesis of β-alanine[J]. Microbiol China, 2013, 40(12):2161-2170. | |
| [4] | 邓思颖, 张君丽, 蔡真, 等. 枯草芽胞杆菌L-天冬氨酸α-脱羧酶的酶学性质[J]. 生物工程学报, 2015, 31(8):1184-1193. |
| Deng SY, Zhang JL, Cai Z, et al. Characterization of L-aspartate-α-decarboxylase from Bacillus subtilis[J]. Chin J Biotechnol, 2015, 31(8):1184-1193. | |
| [5] | 陈涛, 徐世永, 冯炎. 结核杆菌L-天冬氨酸α-脱羧酶诱导表达条件研究[J]. 金陵科技学院学报, 2016, 32(3):80-83. |
| Chen T, Xu SY, Feng Y. Inducing conditions of recombined L-aspartate-α-decarboxylase in fermentor[J]. J Jinling Inst Technol, 2016, 32(3):80-83. | |
| [6] |
Kwon AR, Lee BI, Han BW, et al. Crystallization and preliminary X-ray crystallographic analysis of aspartate 1-decarboxylase from Helicobacter pylori[J]. Acta Crystallogr D Biol Crystallogr, 2002, 58(pt 5):861-863.
doi: 10.1107/S0907444902004833 URL |
| [7] |
Schmitzberger F, Kilkenny ML, Lobley CM, et al. Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase[J]. EMBO J, 2003, 22(23):6193-6204.
pmid: 14633979 |
| [8] | Li HH, Lu XL, Chen KQ, et al. β-alanine production using whole-cell biocatalysts in recombinant Escherichia coli[J]. Mol Catal, 2018, 449:93-98. |
| [9] |
Jang YS, Kim B, Shin JH, et al. Bio-based production of C2-C6 platform chemicals[J]. Biotechnol Bioeng, 2012, 109(10):2437-2459.
doi: 10.1002/bit.24599 URL |
| [10] | 张腾辉. L-天冬氨酸α-脱羧酶的表达、改造及全细胞制备β-丙氨酸[D]. 无锡: 江南大学, 2018. |
| Zhang TH. Expression and modification of L-aspartate α-decarboxylase for the whole-cell transformation of β-alanine[D]. Wuxi: Jiangnan University, 2018. | |
| [11] | 莫芹, 李由然, 石贵阳. 细菌L-天冬氨酸α-脱羧酶的分子机制及分子改造研究进展[J]. 微生物学通报, 2018, 45(7):1546-1554. |
| Mo Q, Li YR, Shi GY. Advances in molecular mechanism and modification of bacterial L-aspartate alpha-decarboxylase[J]. Microbiol China, 2018, 45(7):1546-1554. | |
| [12] | 莫芹. L-天冬氨酸α-脱羧酶催化失活相关分子机制的研究[D]. 无锡: 江南大学, 2019. |
| Mo Q. Molecular mechanism of the catalytic inactivation of L-aspartate alpha-decarboxylase[D]. Wuxi: Jiangnan University, 2019. | |
| [13] | 邢艳珑, 毛相朝, 王舒, 等. 应用荧光分析法检测酶的研究进展[J]. 生物工程学报, 2009, 25(12):1765-1769. |
| Xing YL, Mao N, et al. Recent advances in enzyme assays using fluoremetry[J]. Chin J Biotechnol, 2009, 25(12):1765-1769. | |
| [14] | 刘艳莉, 杨广宇, 王秋岩, 等. 脂肪酶和酯酶的定向进化及其应用[J]. 生物加工过程, 2006, 4(1):16-20, 26. |
| Liu YL, Yang GY, Wang QY, et al. Methods and application of directed evolution of lipase and esterase[J]. Chin J Bioprocess Eng, 2006, 4(1):16-20, 26. | |
| [15] |
Yu XJ, Huang CY, Xu XD, et al. Protein engineering of a pyridoxal-5'-phosphate-dependent l-aspartate-α-decarboxylase from Tribolium castaneum for β-alanine production[J]. Molecules, 2020, 25(6):1280.
doi: 10.3390/molecules25061280 URL |
| [16] | 陈虹. L-天冬氨酸-α-脱羧酶的蛋白质工程改造及其在β-丙氨酸生产中的应用[D]. 杭州: 浙江工业大学, 2019. |
| Chen H. Protein engineering of a L-aspartate-α-decarboxylase and its application in the β-alanine production[D]. Hangzhou: Zhejiang University of Technology, 2019. | |
| [17] | 王鹏. 氨基酸脱羧酶重组表达及应用研究[D]. 南京: 南京大学, 2015. |
| Wang P. Study on recombinant expression and application of amino acid decarboxylase[D]. Nanjing: Nanjing University, 2015. | |
| [18] |
Yu K, Hu S, Huang J, et al. A high-throughput colorimetric assay to measure the activity of glutamate decarboxylase[J]. Enzyme Microb Technol, 2011, 49(3):272-276.
doi: 10.1016/j.enzmictec.2011.06.007 URL |
| [19] |
Jiang H, Xia XX, Feng Y, et al. Development of a robust system for high-throughput colorimetric assay of diverse amino acid decarboxylases[J]. Process Biochem, 2017, 60:27-34.
doi: 10.1016/j.procbio.2017.05.028 URL |
| [20] |
Rosenberg RM, Herreid RM, et al. Indicator assay for amino acid decarboxylases[J]. Anal Biochem, 1989, 181(1):59-65.
pmid: 2817382 |
| [21] |
Gibbons BH, Edsall JT. Rate of hydration of carbon dioxide and dehydration of carbonic acid at 25 degrees[J]. J Biol Chem, 1963, 238:3502-3507.
doi: 10.1016/S0021-9258(18)48696-6 URL |
| [22] |
Persson M, Palcic MM. A high-throughput pH indicator assay for screening glycosyltransferase saturation mutagenesis libraries[J]. Anal Biochem, 2008, 378(1):1-7.
doi: 10.1016/j.ab.2008.03.006 URL |
| [23] |
Martínez-Martínez I, Montoro-García S, Lozada-Ramírez JD, et al. A colorimetric assay for the determination of acetyl xylan esterase or cephalosporin C acetyl esterase activities using 7-amino cephalosporanic acid, cephalosporin C, or acetylated xylan as substrate[J]. Anal Biochem, 2007, 369(2):210-217.
pmid: 17651681 |
| [24] |
Banerjee A, Kaul P, Sharma R, et al. A high-throughput amenable colorimetric assay for enantioselective screening of nitrilase-producing microorganisms using pH sensitive indicators[J]. J Biomol Screen, 2003, 8(5):559-565.
pmid: 14567783 |
| [25] |
Mo Q, Mao A, et al. Substrate inactivation of bacterial L-aspartate α-decarboxylase from Corynebacterium jeikeium K411 and improvement of molecular stability by saturation mutagenesis[J]. World J Microbiol Biotechnol, 2019, 35(4):62.
doi: 10.1007/s11274-019-2629-6 URL |
| [26] |
Pei WL, Zhang JL, Deng SY, et al. Molecular engineering of l-aspartate-α-decarboxylase for improved activity and catalytic stability[J]. Appl Microbiol Biotechnol, 2017, 101(15):6015-6021.
doi: 10.1007/s00253-017-8337-y URL |
| [27] |
Qian Y, Lu C, et al. Engineering protonation conformation of l-aspartate-α-decarboxylase to relieve mechanism-based inactivation[J]. Biotechnol Bioeng, 2020, 117(6):1607-1614.
doi: 10.1002/bit.27316 URL |
| [28] | Chapman E, Wong CH. A pH sensitive colorometric assay for the high-throughput screening of enzyme inhibitors and substrates:a case study using kinases[J]. Bioorg Med Chem, 2002, 10(3):551-555. |
| [29] |
Janes LE, Löwendahl AC, Kazlauskas RJ. Quantitative screening of hydrolase libraries using pH indicators:identifying active and enantioselective hydrolases[J]. Chem Eur J, 1998, 4(11):2324-2331.
doi: 10.1002/(SICI)1521-3765(19981102)4:11<2324::AID-CHEM2324>3.0.CO;2-I URL |
| [30] | 李晓涵, 郝建华, 等. 环糊精葡萄糖基转移酶高效异源表达研究进展[J]. 微生物学通报, 2020, 47(2):615-622. |
| Li XH, Hao JH, et al. Advance in high-level heterologous expression of cyclodextrin glycosyltransferase[J]. Microbiol China, 2020, 47(2):615-622. | |
| [31] | 石增秀, 崔文璟, 周丽, 等. 谷氨酸棒杆菌L-天冬氨酸α-脱羧酶基因的克隆及重组酶性质研究[J]. 生物技术通报, 2013(4):110-115. |
| Shi ZX, Cui WJ, Zhou L, et al. Cloning and Characterization of L-aspartate-α-decarboxylase from Corynebacterium glutamicum[J]. Biotechnol Bull, 2013(4):110-115. | |
| [32] | 田慧, 等. 酰胺酶高通量筛选方法研究进展[J]. 微生物学杂志, 2012, 32(3):66-71. |
| Tian H, et al. Advances in high throughput screening method for amidase[J]. J Microbiol, 2012, 32(3):66-71. |
| [1] | 韩惠, 张舰, 任宇红. 短链脱氢酶Lvchun的分子改造及其在氯霉胺合成中的应用[J]. 生物技术通报, 2023, 39(4): 81-92. |
| [2] | 田庚, 高伟强, 陈晓波, 张春晓. 地衣芽孢杆菌KD-1β-甘露聚糖酶定点突变提高酶活性及稳定性[J]. 生物技术通报, 2021, 37(10): 100-109. |
| [3] | 仲建锋, 李兴奎, 徐重新, 张霄, 刘贤金. Cry1B抗独特型单链抗体的定点突变及生物活性分析[J]. 生物技术通报, 2021, 37(10): 186-195. |
| [4] | 孙熙麟, 蒋振彦, 刘志屹, 戴璐, 孙非, 黄伟. 氨基酸定点突变提高灵芝蛋白LZ-8热稳定性的研究[J]. 生物技术通报, 2020, 36(1): 23-28. |
| [5] | 邱锦, 黄火清, 姚斌, 罗会颖. 解淀粉芽孢杆菌淀粉酶催化活力改良及其在枯草芽孢杆菌中的高效表达[J]. 生物技术通报, 2019, 35(9): 134-143. |
| [6] | 徐林娜, 胡孟可, 童文艳, 李芬. 烟草NtTkr尾部点突变对卷曲螺旋结构及与靶蛋白相互作用的影响[J]. 生物技术通报, 2019, 35(5): 64-69. |
| [7] | 王柳月, 李慧美, 马梦琪, 梁明星, 贺如阳, 陈华波. 利用旁侧引物提高重叠延伸PCR定点突变效率[J]. 生物技术通报, 2019, 35(12): 196-202. |
| [8] | 陈少威, 吴程, 苏月华, 蔡斌斌, 谢盼盼, 杨梅. 苏云金芽胞杆菌aiiA的5'端侧翼序列的克隆与功能鉴定[J]. 生物技术通报, 2018, 34(11): 136-143. |
| [9] | 秦海彬, 熊涛, 张博, 牛坤. α-酮戊二酸半醛脱氢酶的定点突变及酶学性质变化 [J]. 生物技术通报, 2017, 33(8): 180-185. |
| [10] | 曾静, 郭建军, 袁林, 杨罡, 陈俊. 极端嗜热α-淀粉酶ApkA的高温活性和热稳定性的优化研究[J]. 生物技术通报, 2017, 33(8): 192-198. |
| [11] | 邬志杰, 吴更, 唐鸿志, 许平. 恶臭假单胞菌HspB的单晶培养及结晶条件优化[J]. 生物技术通报, 2015, 31(11): 236-242. |
| [12] | 庞浩, 陈燕, 吴倩倩, 刘春宇, 郭媛, 林丽华, 黄日波. 地衣芽孢杆菌(Bacillus licheniformis)外切葡聚糖酶CelB基因的发掘及功能鉴定[J]. 生物技术通报, 2013, 0(9): 151-157. |
| [13] | 段桂华, 沈姗姗, 诸葛宇征. 改良重叠区扩增法构建Rac1相关质粒[J]. 生物技术通报, 2013, 0(6): 177-182. |
| [14] | 樛跃;许崇波;. A型产气荚膜梭菌α毒素Thr-272基因定点突变与结构分析[J]. , 2012, 0(10): 186-192. |
| [15] | 袁野;蔡渊恒;姜世民;姜卫红;. 定点突变提高D-氨甲酰水解酶的可溶性表达[J]. , 2012, 0(01): 94-98. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
