








生物技术通报 ›› 2023, Vol. 39 ›› Issue (1): 214-223.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0388
张君1,2,3( ), 张虹1,2,3, 张芮1,2,3, 路国栋1,2,3, 雍婧姣1,2,3, 郎思睿1,2,3, 陈任1,2,3(
), 张虹1,2,3, 张芮1,2,3, 路国栋1,2,3, 雍婧姣1,2,3, 郎思睿1,2,3, 陈任1,2,3( )
)
收稿日期:2022-04-01
出版日期:2023-01-26
发布日期:2023-02-02
作者简介:张君,女,硕士研究生,研究方向:植物生物学;E-mail: 基金资助:
ZHANG Jun1,2,3( ), ZHANG Hong1,2,3, ZHANG Rui1,2,3, LU Guo-dong1,2,3, YONG Jing-jiao1,2,3, LANG Si-rui1,2,3, CHEN Ren1,2,3(
), ZHANG Hong1,2,3, ZHANG Rui1,2,3, LU Guo-dong1,2,3, YONG Jing-jiao1,2,3, LANG Si-rui1,2,3, CHEN Ren1,2,3( )
)
Received:2022-04-01
Published:2023-01-26
Online:2023-02-02
摘要:
甜叶菊(Stevia rebaudiana Bertoni)生产的甜菊醇糖苷因具有高甜度、低热能、不参与人体内代谢兼具保健功能等特点,被誉为最有发展前途的新糖源。从甜叶菊叶片克隆了甜菊醇糖苷生物合成途径中的关键基因SrUGT85C2、SrUGT91D2m和SrUGT76G1,构建植物基因过量表达载体,以单独或组合的形式将这些基因导入到甜叶菊中,获得转基因植株。与野生型对照植株相比,单独导入SrUGT85C2的转基因植株中甜菊醇单糖苷含量提高,总糖苷、莱包迪苷A含量及占比没有明显变化;单独导入SrUGT91D2m的转基因植株中甜菊醇单糖苷含量显著降低,而甜菊醇双糖苷含量显著增加;单独导入SrUGT76G1的转基因植株中,总糖苷含量显著提高,莱包迪苷A含量达到10%以上,比对照提高了2倍,而甜菊糖苷含量减少了一半。3个基因组合同时导入的转基因甜叶菊植株与单独导入SrUGT76G1的转基因甜叶菊植株类似,其总糖苷、莱包迪苷A含量及其占比均显著提高。这些结果为以后通过分子生物学技术来调控甜菊醇糖苷生物合成关键基因的表达,培育莱包迪苷A含量高的高品质甜叶菊新品系提供了理论依据和技术方法。
张君, 张虹, 张芮, 路国栋, 雍婧姣, 郎思睿, 陈任. 甜菊醇糖苷生物合成关键基因的导入和鉴定分析[J]. 生物技术通报, 2023, 39(1): 214-223.
ZHANG Jun, ZHANG Hong, ZHANG Rui, LU Guo-dong, YONG Jing-jiao, LANG Si-rui, CHEN Ren. Transformation and Functional Identification of the Key Genes Associated with Steviol Glycosides Biosynthesis in Stevia rebaudiana[J]. Biotechnology Bulletin, 2023, 39(1): 214-223.
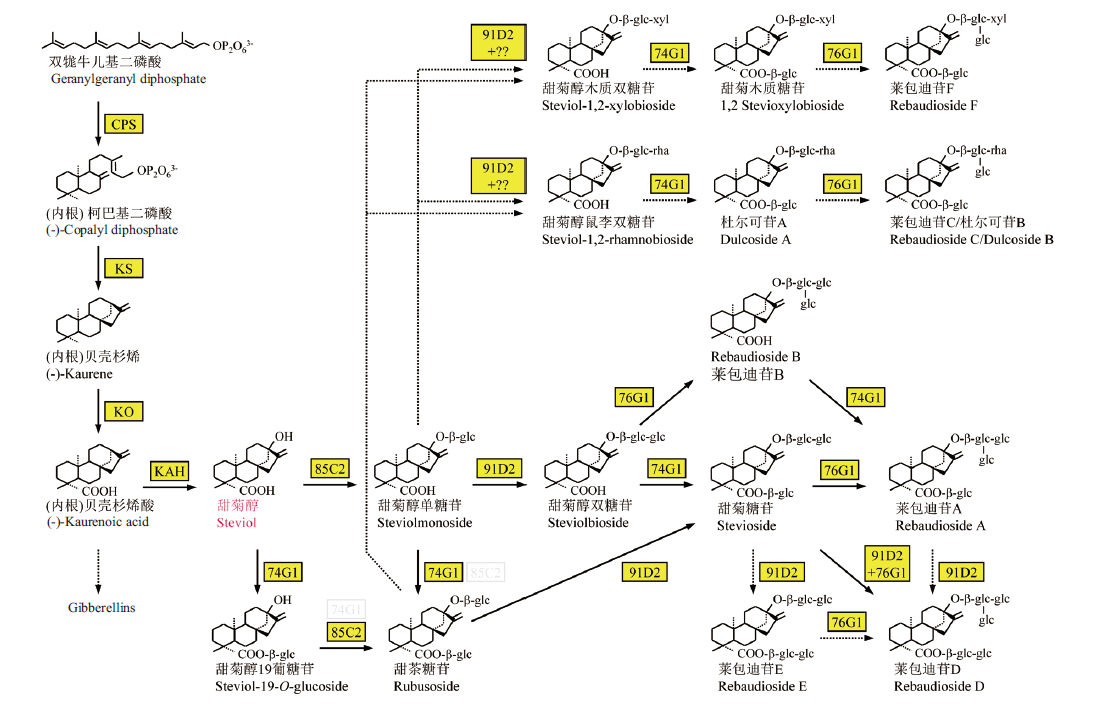
图1 甜菊醇糖苷的生物合成途径 CPS, 柯巴基二磷酸合酶; KS, 贝壳杉烯合酶; KO, 贝壳杉烯氧化酶; KAH, 内根-贝壳杉烯酸羟化酶; 86C2, 91D2, 74G1, 76G1, UDP-葡萄糖基转移酶(UGT86C2, UGT91D2, UGT74G1, UGT76G1)
Fig. 1 Biosynthetic pathways of steviol glycosides CPS, copalylpyrophosphate synthase; KS, kaurene synthase; KO, kaurene oxidized; KAH, kaurenoicacid hydroxylase; 86C2, 91D2, 74G1, 76G1, UDP-glucosyltransferases(UGT86C2, UGT91D2, UGT74G1, UGT76G1)
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引入酶切位点Restriction site |
|---|---|---|
| Sr76G1-S | TATGGATCCTTGCGTGTAAACGTCAGT | BamH I |
| Sr76G1-A | ATGCTCGAGTTATTTACAACGATGAAATGT | Xho I |
| Sr85C2-S | TATGGTACCATGGATGCAATGGCTACAAC | Kpn I |
| Sr85C2-A | GCCGAGCTCCAAAGTTACATCTTAAATAG | Sac I |
| Sr91D2m-S | TTACGGTACCATGGCTACCAGTGACTC | Kpn I |
| Sr91D2m-A | GTGGAGCTCTTAACTCTCATGATCGA | Sac I |
| CR80 MCS insert 35S-S | CACAATCCCACTATCCTTCGCA | |
| CR80 MCS insert NOS-A | TCCACTCTAATCATAAAAACCCATCTC | |
| CR80 MCS insert NOS-S | GAGATGGGTTTTTATGATTAGAGTCC | |
| CR100 Cla-S | CGCTACTGATTACGGTGCTGCTAT | |
| CR100 omega-A | TTGTTTGTTGTTTGTTGTTGTTGGTAA |
表1 PCR扩增引物
Table 1 Primers for PCR amplification
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引入酶切位点Restriction site |
|---|---|---|
| Sr76G1-S | TATGGATCCTTGCGTGTAAACGTCAGT | BamH I |
| Sr76G1-A | ATGCTCGAGTTATTTACAACGATGAAATGT | Xho I |
| Sr85C2-S | TATGGTACCATGGATGCAATGGCTACAAC | Kpn I |
| Sr85C2-A | GCCGAGCTCCAAAGTTACATCTTAAATAG | Sac I |
| Sr91D2m-S | TTACGGTACCATGGCTACCAGTGACTC | Kpn I |
| Sr91D2m-A | GTGGAGCTCTTAACTCTCATGATCGA | Sac I |
| CR80 MCS insert 35S-S | CACAATCCCACTATCCTTCGCA | |
| CR80 MCS insert NOS-A | TCCACTCTAATCATAAAAACCCATCTC | |
| CR80 MCS insert NOS-S | GAGATGGGTTTTTATGATTAGAGTCC | |
| CR100 Cla-S | CGCTACTGATTACGGTGCTGCTAT | |
| CR100 omega-A | TTGTTTGTTGTTTGTTGTTGTTGGTAA |
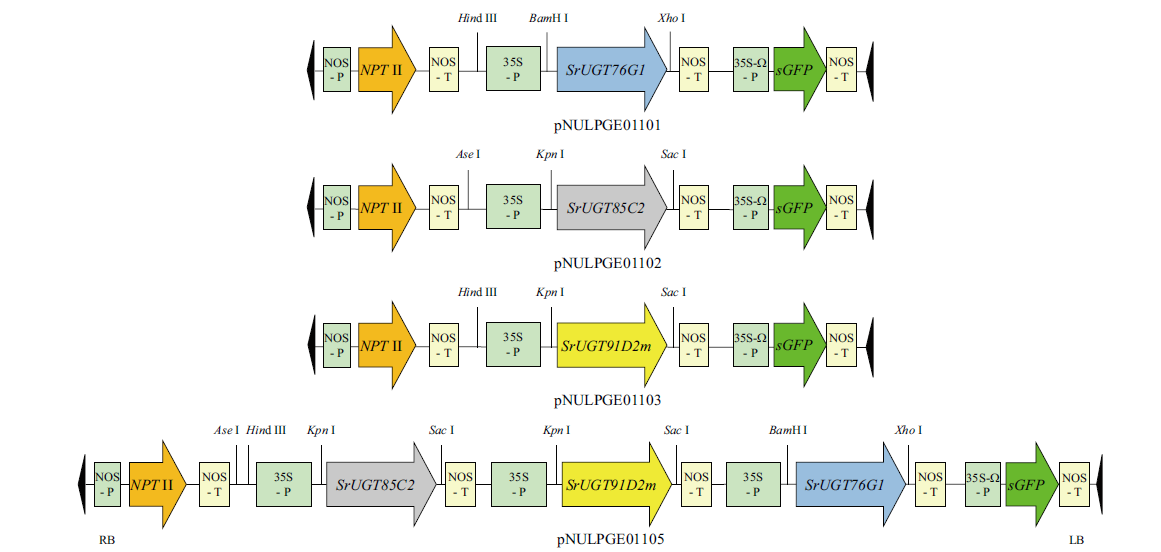
图2 单基因或三基因共表达载体构建示意图 NOS-P,脂碱合酶启动子; NOS-T,脂碱合酶终止子; 35S-P,花椰菜花叶病毒35S启动子; 35S-W-P, 35S启动子附加一个欧米茄翻译增强子元件; NPT II,新霉素磷酸转移酶II; sGFP, 合成的S65T变异绿色荧光蛋白
Fig. 2 Schematic structure of co-expression vectors of single genes or three genes NOS-P, nopaline synthase promoter; NOS-T, nopaline synthase terminator; 35S-P, cauliflower mosaic virus 35S promoter; 35S-W-P, 35S promoter with an additional omega element translational enhancer; NPT II, neomycin phosphotransferase II; sGFP, synthetic green-fluorescent protein with S65T mutation
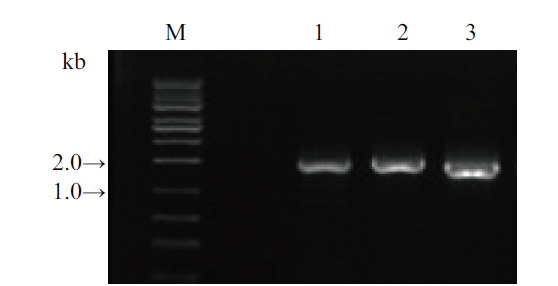
图3 3个基因PCR扩增 1, SrUGT76G1基因(引物Sr76G1-S, A;1 425 bp);2, SrUGT85C2基因(引物Sr85C2-S, A;1 511 bp);3, SrUGT91D2m基因(引物Sr91D2m-S, A;1 441 bp);M, DNA分子量
Fig. 3 PCR amplification of 3 target genes 1, SrUGT76G1(primer pair: Sr76G1-S, A; 1 425 bp); 2, SrUGT85C2(primer pair: Sr85C2-S, A; 1 511 bp); 3, SrUGT91D2m(primer pair: Sr91D2m-S, A; 1 441 bp); M, DNA marker
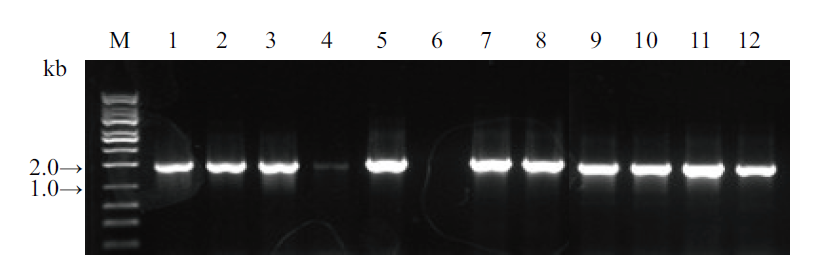
图4 3个基因以单独形式连接至pKAFCR80过渡载体后菌落PCR鉴定 1-4, SrUGT76G1基因组合(1 675 bp); 5-8, SrUGT85C2基因组合(1 764 bp); 9-12, SrUGT91D2m基因组合(1 693 bp)(引物均为CR80 MCS insert 35S-S, A); M, DNA分子量
Fig. 4 Colony PCR identification of the 3 genes individually constructed into pKAFCR80 intermediate vector 1-4, SrUGT76G1(1 675 bp); 5-8, SrUGT85C2(1 764 bp); 9-12, SrUGT91D2m(1 693 bp)(All primer pair: CR80 MCS insert 35S-S, A); M, DNA marker
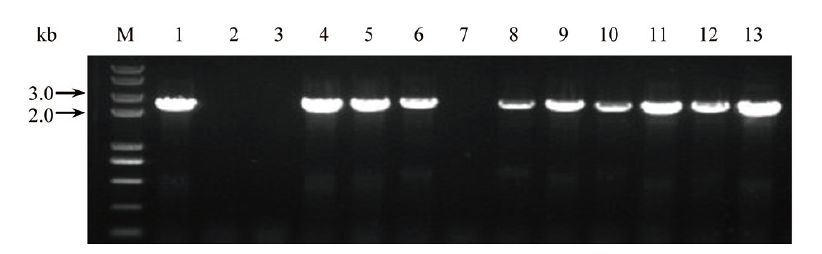
图5 3个基因以单独形式连接至pKAFCR100二元表达载体后菌落PCR鉴定 1-5, SrUGT76G1基因组合(2 901 bp); 6-9, SrUGT85C2基因组合(2 808 bp); 10-13, SrUGT91D2m基因组合(2 919 bp)(引物均为CR100 Cla-S, CR80 MCS insert NOS-A); M, DNA分子量
Fig. 5 Colony PCR identification of the 3 genes individu-ally constructed into pKAFCR100 binary vector 1-5, SrUGT76G1(2 901 bp); 6-9, SrUGT85C2(2 808 bp); 10-13, SrUGT91D2m(2 919 bp)(All primers pair: CR100 Cla-S, CR80 MCS insert NOS-A); M, DNA maker
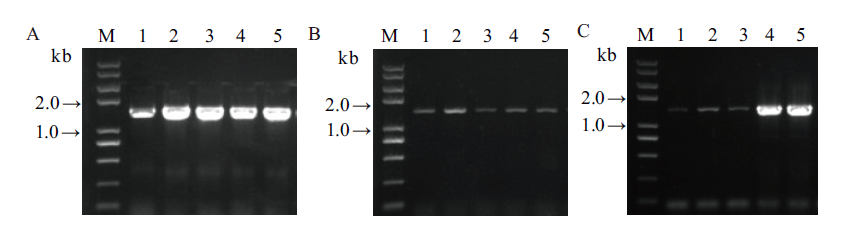
图6 3个基因以组合形式连接至pKAFCR100二元表达载体菌落PCR鉴定 A: 1-5, SrUGT76G1基因(引物Sr76G1-S, A; 1 425 bp); B: 1-5, SrUGT85C2基因(引物Sr85C2-S, A; 1 511 bp); C: 1-5, SrUGT91D2m基因(引物Sr91D2m-S, A; 1 441 bp); M, DNA 分子量
Fig. 6 Colony PCR identification of 3 genes in combination constructed into pKAFCR100 binary vector A: 1-5, SrUGT76G1(primer pair: Sr76G1-S, A; 1 425 bp); B: 1-5, SrUGT85C2(primer pair: Sr85C2-S, A; 1 511 bp); C: 1-5, SrUGT91D2m(primer pair: Sr91D2m-S, A; 1 441 bp); M, DNA marker
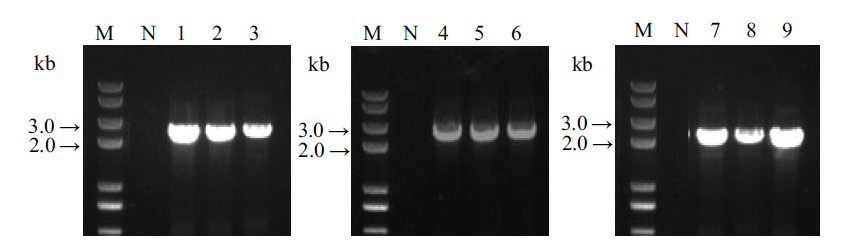
图7 3个基因以单独形式转化植株的PCR鉴定 N, 野生型阴性对照, 1-3, SrUGT76G1基因单独导入(引物Sr76G1-S, CR100 omega-A; 2 151 bp); 4-6, SrUGT85C2基因单独导入(引物CR100 Cla-S, Sr85C2-A; 2 643 bp); 7-9, SrUGT91D2m基因单独导入(引物CR80 MCS insert NOS-S, Sr91D2m-A; 2 434 bp); M, DNA 分子量
Fig. 7 PCR identification of the transgenic plant transferr-ed 3 genes individually N, wild-type negative control, 1-3, SrUGT76G1 gene transferred individually(primer pair: Sr76G1-S, CR100 omega-A; 2 151 bp); 4-6, SrUGT85C2 gene transferred individually(primer pair: CR100 Cla-S, Sr85C2-A; 2 643 bp); 7-9, SrUGT91D2m gene transferred individually(primer pair: CR80 MCS insert NOS-S, Sr91D2m-A; 2 434 bp); M, DNA marker
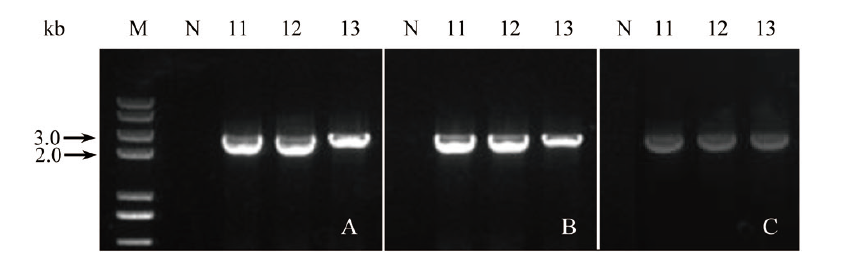
图8 3个基因以组合形式转化植株的PCR鉴定 A: N, 野生型阴性对照, 11-13, 3个基因以组合形式导入(引物Sr76G1-S, pKAFCR100 omega-A; 2 151 bp); B: N, 野生型阴性对照, 11-13, 3个基因以组合形式导入(引物pKAFCR100 Cla-S, Sr85C2-A; 2 643 bp); C: N, 野生型阴性对照, 11-13, 3个基因以组合形式导入(引物CR80 MCS insert NOS-S, Sr91D2m-A; 2 434 bp); M, DNA 分子量
Fig. 8 PCR identification of the transgenic plant transferr-ed 3 genes in combination A: N, wild-type negative control, 11-13, three genes were transferred in combination(primer pair: Sr76G1-S, pKAFCR100 omega-A; 2 151 bp). B: N, wild-type negative control, 11-13, three genes were transferred in combination(primer pair: pKAFCR100 Cla-S, Sr85C2-A; 2 643 bp). C: N, wild-type negative control, 11-13, three genes were transferred in combination(primer pair: CR80 MCS insert NOS-S, Sr91D2m-A; 2 434 bp). M, DNA marker
| 转基因植株Transgenic plant | SGs/% | SMono/% | SBio/% | ST/% | RA/% | RA/SGs/% |
|---|---|---|---|---|---|---|
| 野生型对照Wild-type control | 12.74 ± 1.17c | 1.26 ± 0.13b | 0.66 ± 0.07c | 5.47 ± 0.65b | 5.34 ± 0.52c | 40.56 ± 2.36c |
| Sr01102(SrUGT85C2) | 13.22 ± 1.46bc | 1.57 ± 0.17a | 0.74 ± 0.06b | 5.55 ± 0.84b | 5.36 ± 0.64c | 40.79 ± 2.20c |
| Sr01103(SrUGT91D2m) | 13.65 ± 1.16b | 0.63 ± 0.07c | 1.10 ± 0.15a | 5.98 ± 0.56a | 5.95 ± 0.64c | 42.79 ± 2.73c |
| Sr01101(SrUGT76G1) | 15.22 ± 1.25ab | 1.22 ± 0.12b | 0.74 ± 0.06b | 2.49 ± 0.24d | 10.77 ± 1.02a | 71.70 ± 2.43a |
| Sr01105(3个基因组合/3 genes Combined) | 16.12 ± 1.59a | 1.23 ± 0.11b | 0.83 ± 0.09b | 4.60 ± 0.36c | 9.45 ± 1.15b | 58.27 ± 1.96b |
表2 不同转基因植株中各种甜菊醇糖苷的含量
Table 2 Variety of steviol glycosides contents in different transgenic plant
| 转基因植株Transgenic plant | SGs/% | SMono/% | SBio/% | ST/% | RA/% | RA/SGs/% |
|---|---|---|---|---|---|---|
| 野生型对照Wild-type control | 12.74 ± 1.17c | 1.26 ± 0.13b | 0.66 ± 0.07c | 5.47 ± 0.65b | 5.34 ± 0.52c | 40.56 ± 2.36c |
| Sr01102(SrUGT85C2) | 13.22 ± 1.46bc | 1.57 ± 0.17a | 0.74 ± 0.06b | 5.55 ± 0.84b | 5.36 ± 0.64c | 40.79 ± 2.20c |
| Sr01103(SrUGT91D2m) | 13.65 ± 1.16b | 0.63 ± 0.07c | 1.10 ± 0.15a | 5.98 ± 0.56a | 5.95 ± 0.64c | 42.79 ± 2.73c |
| Sr01101(SrUGT76G1) | 15.22 ± 1.25ab | 1.22 ± 0.12b | 0.74 ± 0.06b | 2.49 ± 0.24d | 10.77 ± 1.02a | 71.70 ± 2.43a |
| Sr01105(3个基因组合/3 genes Combined) | 16.12 ± 1.59a | 1.23 ± 0.11b | 0.83 ± 0.09b | 4.60 ± 0.36c | 9.45 ± 1.15b | 58.27 ± 1.96b |
| [1] |
Soejarto DD, Kinghorn AD, Farnsworth NR. Potential sweetening agents of plant origin. III. Organoleptic evaluation of Stevia leaf herbarium samples for sweetness[J]. J Nat Prod, 1982, 45(5): 590-599.
pmid: 7153776 |
| [2] |
Kinghorn AD, Soejarto DD, Nanayakkara NPD, et al. A phytochemical screening procedure for sweet ent-kaurene glycosides in the genus Stevia[J]. J Nat Prod, 1984, 47(3): 439-444.
pmid: 6481357 |
| [3] |
Goyal SK, Samsher, Goyal RK. Stevia(Stevia rebaudiana)a bio-sweetener: a review[J]. Int J Food Sci Nutr, 2010, 61(1): 1-10.
doi: 10.3109/09637480903193049 pmid: 19961353 |
| [4] |
Yadav AK. A review on the improvement of Stevia[Stevia rebaudiana(Bertoni)[J]. Can J Plant Sci, 2011, 91(1): 1-27.
doi: 10.4141/cjps10086 URL |
| [5] | 倪军明, 李军平. 甜菊糖工业发展现状与前景[J]. 广州食品工业科技, 2004, 20(3): 156-158. |
| Ni JM, Li JP. Current development and prospect of stevioside industry[J]. Guangzhou Food Sci Technol, 2004, 20(3): 156-158. | |
| [6] | 徐鸿. 甜菊糖的特点及功能[J]. 现代农业科技, 2013(2): 286-287. |
| Xu H. Characteristic and functions of stevioside[J]. Mod Agric Sci Technol, 2013(2): 286-287. | |
| [7] |
Ohta M, Sasa S, Inoue A, et al. Characterization of novel steviol glycosides from leaves of Stevia rebaudiana morita[J]. J Appl Glycosci, 2010, 57(3): 199-209.
doi: 10.5458/jag.57.199 URL |
| [8] |
Prakash I, Campbell M, Chaturvedula VSP. Catalytic hydrogenation of the sweet principles of Stevia rebaudiana, Rebaudioside B, Rebaudioside C, and Rebaudioside D and sensory evaluation of their reduced derivatives[J]. Int J Mol Sci, 2012, 13(11): 15126-15136.
doi: 10.3390/ijms131115126 pmid: 23203115 |
| [9] |
Wölwer-Rieck U. The leaves of Stevia rebaudiana(Bertoni), their constituents and the analyses thereof: a review[J]. J Agric Food Chem, 2012, 60(4): 886-895.
doi: 10.1021/jf2044907 URL |
| [10] |
Tada A, Takahashi K, Ishizuki K, et al. Absolute quantitation of stevioside and rebaudioside A in commercial standards by quantitative NMR[J]. Chem Pharm Bull(Tokyo), 2013, 61(1): 33-38.
pmid: 23124594 |
| [11] | Crammer B, Ikan R. Sweet glycosides from the stevia plant[J]. Chemistry in Britain, 1986, 22(10): 915-916. |
| [12] | 孙传范, 李进伟. 甜菊糖苷研究进展[J]. 食品科学, 2010, 31(9): 338-340. |
| Sun CF, Li JW. Research progress on steviosides[J]. Food Sci, 2010, 31(9): 338-340. | |
| [13] | Persinos G. Method of producing stevioside: US3723410[P]. 1973-03-27. |
| [14] |
Humphrey TV, Richman AS, Menassa R, et al. Spatial organisation of four enzymes from Stevia rebaudiana that are involved in steviol glycoside synthesis[J]. Plant Mol Biol, 2006, 61(1/2): 47-62.
doi: 10.1007/s11103-005-5966-9 URL |
| [15] |
Brandle JE, Telmer PG. Steviol glycoside biosynthesis[J]. Phytochemistry, 2007, 68(14): 1855-1863.
pmid: 17397883 |
| [16] |
Mohamed AAA, Ceunen S, Geuns JMC, et al. UDP-dependent glycosyltransferases involved in the biosynthesis of steviol glycosides[J]. J Plant Physiol, 2011, 168(10): 1136-1141.
doi: 10.1016/j.jplph.2011.01.030 URL |
| [17] |
Ceunen S, Geuns JMC. Steviol glycosides: chemical diversity, metabolism, and function[J]. J Nat Prod, 2013, 76(6): 1201-1228.
doi: 10.1021/np400203b pmid: 23713723 |
| [18] |
Yadav SK, Guleria P. Steviol glycosides from Stevia: biosynthesis pathway review and their application in foods and medicine[J]. Crit Rev Food Sci Nutr, 2012, 52(11): 988-998.
doi: 10.1080/10408398.2010.519447 URL |
| [19] |
Guleria P, Yadav SK. Agrobacterium mediated transient gene silencing(AMTS)in Stevia rebaudiana: insights into steviol glycoside biosynthesis pathway[J]. PLoS One, 2013, 8(9): e74731.
doi: 10.1371/journal.pone.0074731 URL |
| [20] |
Modi A, Litoriya N, Prajapati V, et al. Transcriptional profiling of genes involved in steviol glycoside biosynthesis in Stevia rebaudiana bertoni during plant hardening[J]. Dev Dyn, 2014, 243(9): 1067-1073.
doi: 10.1002/dvdy.24157 URL |
| [21] |
Madhav H, Bhasker S, Chinnamma M. Functional and structural variation of uridine diphosphate glycosyltransferase(UGT)gene of Stevia rebaudiana-UGTSr involved in the synthesis of rebaudioside A[J]. Plant Physiol Biochem, 2013, 63: 245-253.
doi: 10.1016/j.plaphy.2012.11.029 URL |
| [22] |
Guleria P, Masand S, Yadav SK. Overexpression of SrUGT85C2 from Stevia reduced growth and yield of transgenic Arabidopsis by influencing plastidial MEP pathway[J]. Gene, 2014, 539(2): 250-257.
doi: 10.1016/j.gene.2014.01.071 URL |
| [23] |
Yang YH, Huang SZ, Han YL, et al. Base substitution mutations in uridinediphosphate-dependent glycosyltransferase 76G1 gene of Stevia rebaudiana causes the low levels of rebaudioside A: mutations in UGT76G1, a key gene of steviol glycosides synthesis[J]. Plant Physiol Biochem, 2014, 80: 220-225.
doi: 10.1016/j.plaphy.2014.04.005 URL |
| [24] |
Kim MJ, Zheng JS, et al. Overexpression of SrUGT76G1 in Stevia alters major steviol glycosides composition towards improved quality[J]. Plant Biotechnol J, 2019, 17(6): 1037-1047.
doi: 10.1111/pbi.13035 URL |
| [25] | 陈任, 张虹, 张芮, 等. 用于植物多基因表达载体构建的质粒系统[J]. 分子植物育种, 2018, 16(4): 1138-1146. |
| Chen R, Zhang H, Zhang R, et al. A plasmid system for construction of plant multiple gene expression vectors[J]. Mol Plant Breed, 2018, 16(4): 1138-1146. | |
| [26] | 张芮. 甜菊醇糖苷生物合成途径关键基因的功能评价[D]. 银川: 宁夏大学, 2016. |
| Zhang R. The functional evaluation of key genes in steviol glycosides biosynthetic pathway[D]. Yinchuan: Ningxia University, 2016. | |
| [27] | 张虹, 张芮, 薄玉瑶, 等. 甜叶菊甜菊醇糖苷生物合成关键酶基因表达量与莱宝迪苷A含量的相关关系[J]. 分子植物育种, 2015, 13(8): 1802-1807. |
| Zhang H, Zhang R, Bo YY, et al. Correlation between the transcript levels of key enzyme-encoding genes and the content of rebaudioside a in steviol glycoside biosynthesis of Stevia rebaudiana[J]. Mol Plant Breed, 2015, 13(8): 1802-1807. | |
| [28] |
Jentzer JB, Alignan M, Vaca-Garcia C, et al. Response surface methodology to optimise accelerated solvent extraction of steviol glycosides from Stevia rebaudiana Bertoni leaves[J]. Food Chem, 2015, 166: 561-567.
doi: 10.1016/j.foodchem.2014.06.078 URL |
| [29] |
Periche A, Castelló ML, Heredia A, et al. Influence of drying method on steviol glycosides and antioxidants in Stevia rebaudiana leaves[J]. Food Chem, 2015, 172: 1-6.
doi: 10.1016/j.foodchem.2014.09.029 pmid: 25442516 |
| [30] |
Chranioti C, Chanioti S, Tzia C. Comparison of spray, freeze and oven drying as a means of reducing bitter aftertaste of steviol glycosides(derived from Stevia rebaudiana Bertoni plant)—Evaluation of the final products[J]. Food Chem, 2016, 190: 1151-1158.
doi: 10.1016/j.foodchem.2015.06.083 URL |
| [31] |
张虹, 路国栋, 袁春春, 等. 甜叶菊中9种甜菊醇糖苷积累与其生物合成关键基因表达量的相关性[J]. 核农学报, 2022, 36(1): 75-82.
doi: 10.11869/j.issn.100-8551.2022.01.0075 |
| Zhang H, Lu GD, Yuan CC, et al. Correlation between the accumulations of 9 steviol glycosides and the expressions of the key genes involved in their biosynthesis in Stevia rebaudiana[J]. J Nucl Agric Sci, 2022, 36(1): 75-82. | |
| [32] |
Zhang SS, Yang YS, Lyu CC, et al. Identification of the key residues of the uridine diphosphate glycosyltransferase 91D2 and its effect on the accumulation of steviol glycosides in Stevia rebaudiana[J]. J Agric Food Chem, 2021, 69(6): 1852-1863.
doi: 10.1021/acs.jafc.0c07066 URL |
| [33] | 刘欢, 李艳, 等. 甜叶菊糖基转移酶UGT76G1的克隆表达及其性质研究[J]. 食品工业科技, 2012, 33(20): 187-190. |
| Liu H, Li Y, et al. Cloning, expression and characteristic of glycosyl-transferase UGT76G1 from Stevia rebaudiana[J]. Sci Technol Food Ind, 2012, 33(20): 187-190. | |
| [34] |
Saifi M, Yogindran S, et al. Co-expression of anti-miR319g and miRStv_11 lead to enhanced steviol glycosides content in Stevia rebaudiana[J]. BMC Plant Biol, 2019, 19(1): 274.
doi: 10.1186/s12870-019-1871-2 URL |
| [35] |
Zhang T, Xu XY, Sun YM, et al. The SrWRKY71 transcription factor negatively regulates SrUGT76G1 expression in Stevia rebau-diana[J]. Plant Physiol Biochem, 2020, 148: 26-34.
doi: 10.1016/j.plaphy.2020.01.001 URL |
| [1] | 冯翠莲, 张树珍. 抗虫转基因甘蔗的培育及其抗性丧失的防控策略[J]. 生物技术通报, 2020, 36(7): 209-219. |
| [2] | 朱静雯, 郭书巧, 束红梅, 巩元勇, 蒋璐, 倪万潮. 甜叶菊中新KAH基因的克隆及表达模式分析[J]. 生物技术通报, 2017, 33(4): 104-107. |
| [3] | 余波颖, 王宁琳, 李国婧, 夏亦荠. 基因工程和代谢工程在甜菊糖生产上应用进展[J]. 生物技术通报, 2015, 31(9): 8-14. |
| [4] | 王琼, 刘伯斌, 孔倩倩, 陈介南. 能源植物麻疯树分子育种研究进展[J]. 生物技术通报, 2014, 0(3): 1-8. |
| [5] | 阳辛凤;徐林;弓淑芳;郭安平;孔华;贺立卡;. CUP1为筛选标记的酿酒酵母整合型多基因表达载体的构建[J]. , 2011, 0(10): 139-144. |
| [6] | 倪万潮;郭书巧;. 甜菊醇糖苷生物合成及关键酶研究进展[J]. , 2008, 0(02): 48-53. |
| [7] | 杨奇志;赵琦;孔建强;李高岩. 基因枪技术在农作物基因转化中的应用和进展[J]. , 2003, 0(06): 14-17. |
| [8] | 王景雪;孙毅;杜建中. 玉米转基因研究进展[J]. , 2001, 0(02): 13-16. |
| [9] | 杨淑培. 我国培育成功抗卷叶病毒转基因马铃薯[J]. , 2001, 0(02): 51-51. |
| [10] | Rafael Rangel-aldao;汪开治. 新的国际农业生物技术合作在北京起步[J]. , 2000, 0(05): 37-38. |
| [11] | 王瑶;林木兰;沈锡辉;柳晟. 农杆菌介导的木本植物遗传转化[J]. , 1999, 0(06): 23-27. |
| [12] | . 近期文献集锦[J]. , 1997, 0(06): 52-53. |
| [13] | 朱遐. 碳化硅纤维介导的转化[J]. , 1996, 0(02): 13-13. |
| [14] | 钟蓉;罗鹏;梁明山;刘玉乐. 植物杂种优势利用的基因工程研究进展[J]. , 1996, 0(01): 1-5. |
| [15] | 李思经. 反义基因促进芥菜形态发生[J]. , 1996, 0(01): 11-11. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||