








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 206-217.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0532
平怀磊1( ), 郭雪2, 余潇2, 宋静2, 杜春1, 王娟3(
), 郭雪2, 余潇2, 宋静2, 杜春1, 王娟3( ), 张怀璧4
), 张怀璧4
收稿日期:2022-04-29
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
王娟,女,博士,教授,博士生导师,研究方向:生物多样性保护、植物学、生态学;E-mail:Schima@163.com作者简介:平怀磊,男,硕士研究生,研究方向:生物多样性保护与利用;E-mail:2681418377@qq.com
基金资助:
PING Huai-lei1( ), GUO Xue2, YU Xiao2, SONG Jing2, DU Chun1, WANG Juan3(
), GUO Xue2, YU Xiao2, SONG Jing2, DU Chun1, WANG Juan3( ), ZHANG Huai-bi4
), ZHANG Huai-bi4
Received:2022-04-29
Published:2023-03-26
Online:2023-04-10
摘要:
花色是观赏植物最重要的品质性状之一,在植物生长发育过程中发挥着重要作用。探明滇牡丹的花色调控机理,为提高观赏价值奠定理论基础。以花瓣为材料,克隆获得PdANS,生物信息学分析其特征,结合实时荧光定量PCR、HPLC等技术探究不同组织、不同发育时期中PdANS的表达情况与各组织花青素含量的相关性。结果表明,PdANS全长1 121 bp,包含一个1 064 bp的开放阅读框(ORF),编码354个氨基酸,相对分子质量40.41 kD,理论等电点(pI)5.48,分子式为C1819H2876N474O540S12,脂肪指数为88.64,不稳定指数(II)为55.32,亲水平均数为-0.435,推测为不稳定的亲水蛋白。且无信号肽序列及跨膜螺旋区,是没有分泌功能的非跨膜蛋白。系统进化分析显示,滇牡丹PdANS与牡丹、芍药等植物的ANS蛋白亲缘关系最近。RT-qPCR结果显示,PdANS在花瓣、花药、萼片、苞片和花梗中均有表达,以花瓣中的表达量最高。HPLC结果表明,在不同组织提取液中,分别检测出Cy3G5G、Pn3G5G、Cy3G和Pn3G四种花青素,且花青素含量花瓣>花药>萼片>花梗>苞片。相关性分析表明,花青素含量与PdANS表达量呈极显著相关性。推测PdANS在滇牡丹花色的形成中扮演着重要角色,参与花青素物质的生物合成。
平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217.
PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content[J]. Biotechnology Bulletin, 2023, 39(3): 206-217.

图1 滇牡丹各时期的划分阶段 A-1:滇牡丹红花(S1-S3);B-1:滇牡丹紫花(S1-S3);C-1:滇牡丹红斑黄花(S1-S3)
Fig. 1 The division of different periods of Paeonia delavayi A-1: Red flower of Paeonia delavayi(S1-S3). B-1: Purple flower of Paeonia delavayi(S1-S3). C-1: Yellow flower with red spots of Paeonia delavayi(S1-S3)
| 引物名称Primer name | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| PdANS-ORF | AAACAGAGAGTAAACGTTGCTG | TATAAATCCATCCCGGCCCA |
| PdANS-RT | ATCCTCCACAACATGGTGCC | TCACCAATCCCCTGTGAAGA |
| GAPDH-RT | CACCACTAACTGTCTTGCCC | GCTGCTGGGAATAATGTTG |
表1 PdANS的克隆和qPCR引物分析
Table 1 Primers for PdANS gene cloning and qPCR analysis
| 引物名称Primer name | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| PdANS-ORF | AAACAGAGAGTAAACGTTGCTG | TATAAATCCATCCCGGCCCA |
| PdANS-RT | ATCCTCCACAACATGGTGCC | TCACCAATCCCCTGTGAAGA |
| GAPDH-RT | CACCACTAACTGTCTTGCCC | GCTGCTGGGAATAATGTTG |
图2 滇牡丹RNA琼脂糖凝胶电泳检测 A:滇牡丹花瓣RNA;B:紫花其余组织RNA;M:BM2000+ DNA marker
Fig. 2 P. delavayi RNA agarose gel electrophoresis detection A: Petal RNA of P. delavayi. B: RNA from other tissues of purple flower. M: BM2000+ DNA marker

图3 PdANS全长cDNA的克隆 1-3:滇牡丹PdANS的PCR扩增产物;M:BM2000+ DNA marker
Fig. 3 Cloning of full length cDNA of PdANS gene 1-3: PCR products of PdANS. M: BM2000+ DNA marker

图8 PdANS与其他植物ANS氨基酸序列的多重比对分析 PdANS:滇牡丹;PlANS1:黄牡丹,ALX36000.1;PsANS:牡丹,AIU98515.1;PlANS:芍药,QIC54082.1;TcANS:可可,XP_007040068.2;DzANS:榴莲,XP_022732774.1;CoANS:长蒴黄麻,OMO81016.1;VaANS:野葡萄,ACN38270.1;NgANS:大齿牛果藤,AGO02175.1。下划线表示2OG-Fe II_Oxy蛋白功能域
Fig. 8 Multiple alignment and analysis of amino acid sequence PdANS with ANS from other plants PdANS: Paeonia delavayi. PlANS1: Paeonia lutea, ALX36000.1. PsANS: Paeonia suffruticosa, AIU98515.1. PlANS: Paeonia lactiflora, QIC54082.1. TcANS: Theobroma cacao, XP_007040068.2. DzANS: Durio zibethinus, XP_022732774.1. CoANS: Corchorus olitorius, OMO81016.1. VaANS: Vitis amurensis, ACN38270.1. NgANS: Nekemias grossedentata, AGO02175.1. The underscore indicates the 2OG-Fe II_Oxy protein domain
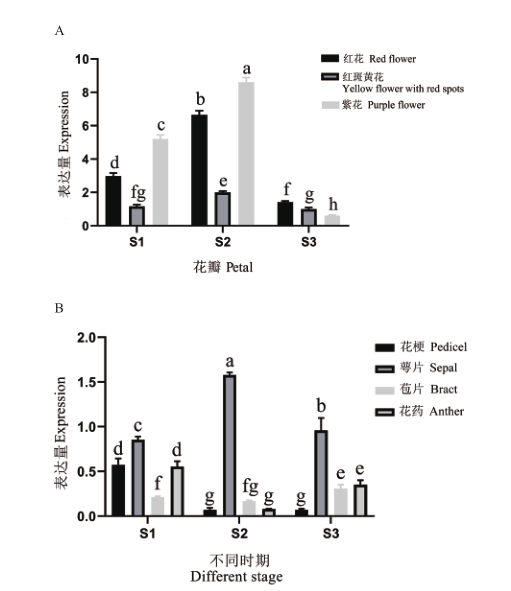
图10 PdANS的表达模式 A:花瓣中的表达模式;B:紫花滇牡丹其余组织的表达模式。S1:硬蕾期;S2:透色期;S3:盛开期。不同小写字母表示显著差异(P<0.05)。下同
Fig. 10 Expression pattern of PdANS A: Patterns of expression in petals. B: Expression patterns in other tissues of the purple flower Paeonia delavayi. S1: Unpigmented tight bud. S2: Slightly pigmented bud. S3: Fully opened flower. Different lowercase letters indicate significant difference(P<0.05). The same below
| 标准品 Standard | 标准曲线 Calibration curve | R2 |
|---|---|---|
| 矢车菊素-3,5二葡萄糖苷(Gy3G5G) | y1=0.925 4x-1.247 1 | 0.999 8 |
| 芍药素-3,5二葡萄糖苷(Pn3G5G) | y2 =1.238 4x-59.599 | 0.999 3 |
| 矢车菊素-3-O-葡萄糖苷(Cy3G) | y3 =5.795x-77.033 | 0.999 6 |
| 芍药素-3-O-葡萄糖苷(Pn3G) | y4 =4.831 5x-12.906 | 0.999 9 |
表2 4种花青素的回归方程
Table 2 Regression equations of four anthocyanins
| 标准品 Standard | 标准曲线 Calibration curve | R2 |
|---|---|---|
| 矢车菊素-3,5二葡萄糖苷(Gy3G5G) | y1=0.925 4x-1.247 1 | 0.999 8 |
| 芍药素-3,5二葡萄糖苷(Pn3G5G) | y2 =1.238 4x-59.599 | 0.999 3 |
| 矢车菊素-3-O-葡萄糖苷(Cy3G) | y3 =5.795x-77.033 | 0.999 6 |
| 芍药素-3-O-葡萄糖苷(Pn3G) | y4 =4.831 5x-12.906 | 0.999 9 |
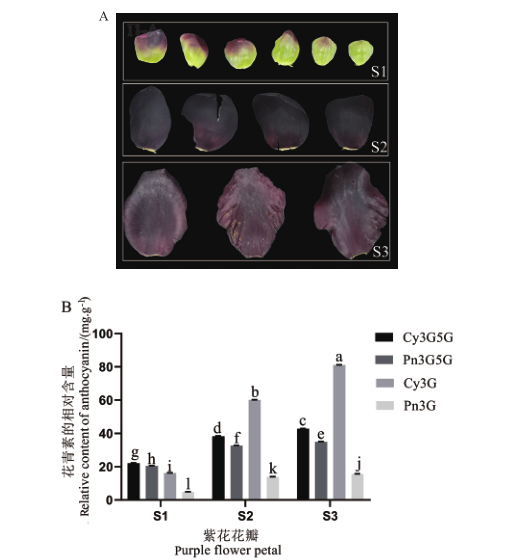
图11 紫花花瓣的花青素分布(A)及含量(B)分析 Cy3G5G:矢车菊素-3,5二葡萄糖苷;Pn3G5G:芍药素-3,5二葡萄糖苷;Cy3G:矢车菊素-3-O-葡萄糖苷;Pn3G:芍药素-3-O-葡萄糖苷。下同
Fig. 11 Analysis of anthocyanin distribution(A)and content(B)in purple flower petals Cy3G5G: Cyanidin-3,5-diglucoside. Pn3G5G: Peonidin-3,5-diglucoside. Cy3G: Cyanidin-3-O-glucoside. Pn3G: Peonidin-3-O-glucoside. The same below
| 项目 Item | PdANS | 花青素含量 Anthocyanin content |
|---|---|---|
| PdANS | 1 | 0.436** |
| 花青素含量 Anthocyanin content | 0.436** | 1 |
表3 花青素含量与PdANS表达量的相关性分析
Table 3 Correlation analysis between anthocyanin content and PdANS gene expression
| 项目 Item | PdANS | 花青素含量 Anthocyanin content |
|---|---|---|
| PdANS | 1 | 0.436** |
| 花青素含量 Anthocyanin content | 0.436** | 1 |
| [1] | 侯夫云, 王庆美, 李爱贤, 等. 植物花青素合成酶的研究进展[J]. 中国农学通报, 2009, 25(21): 188-190. |
|
Hou FY, Wang QM, Li AX, et al. Study progress on anthocyanidin synthase of plants[J]. Chin Agric Sci Bull, 2009, 25(21): 188-190.
doi: 10.11924/j.issn.1000-6850.2009-1169 |
|
| [2] | 赵启明, 李范, 李萍. 花青素生物合成关键酶的研究进展[J]. 生物技术通报, 2012(12): 25-32. |
| Zhao QM, Li F, Li P. Research advances on core enzymes of anthocyanidin biosynthesis[J]. Biotechnol Bull, 2012(12): 25-32. | |
| [3] | Lev-Yadun S, Gould KS. Role of anthocyanins in plant defence[M]//Anthocyanins. New York: Springer New York, 2008: 22-28. |
| [4] |
Butelli E, Titta L, Giorgio M, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors[J]. Nat Biotechnol, 2008, 26(11): 1301-1308.
doi: 10.1038/nbt.1506 pmid: 18953354 |
| [5] |
Sun CD, Huang HZ, Xu CJ, et al. Biological activities of extracts from Chinese bayberry(Myrica rubra Sieb. et Zucc.): a review[J]. Plant Foods Hum Nutr, 2013, 68(2): 97-106.
doi: 10.1007/s11130-013-0349-x URL |
| [6] | 贾赵东, 马佩勇, 边小峰, 等. 植物花青素合成代谢途径及其分子调控[J]. 西北植物学报, 2014, 34(7): 1496-1506. |
| Jia ZD, Ma PY, Bian XF, et al. Biosynthesis metabolic pathway and molecular regulation of plants anthocyanin[J]. Acta Bot Boreali Occidentalia Sin, 2014, 34(7): 1496-1506. | |
| [7] | 魏永赞, 李伟才, 董晨, 等. 光照对植物花色素苷生物合成的调控及机制[J]. 植物生理学报, 2017, 53(9): 1577-1585. |
| Wei YZ, Li WC, Dong C, et al. Regulation and mechanism of light on anthocyanin biosynthesis in plants[J]. Plant Physiol J, 2017, 53(9): 1577-1585. | |
| [8] |
Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits[J]. Trends Plant Sci, 2013, 18(9): 477-483.
doi: 10.1016/j.tplants.2013.06.003 pmid: 23870661 |
| [9] |
Pelletier MK, Murrell JR, Shirley BW. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of “early” and “late” genes[J]. Plant Physiol, 1997, 113(4): 1437-1445.
pmid: 9112784 |
| [10] |
Cheng LQ, Xu YJ, Grotewold E, et al. Characterization of anthocyanidin synthase(ANS)gene and anthocyanidin in rare medicinal plant-Saussurea medusa[J]. Plant Cell Tissue Organ Cult, 2007, 89(1): 63-73.
doi: 10.1007/s11240-007-9211-x URL |
| [11] |
Li XG, Wang J, Yu ZY. Cloning of an anthocyanidin synthase gene homolog from blackcurrant(Ribes nigrum L.)and its expression at different fruit stages[J]. Genet Mol Res, 2015, 14(1): 2726-2734.
doi: 10.4238/2015.March.31.2 pmid: 25867421 |
| [12] |
Sparvoli F, Martin C, Scienza A, et al. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape(Vitis vinifera L.)[J]. Plant Mol Biol, 1994, 24(5): 743-755.
doi: 10.1007/BF00029856 pmid: 8193299 |
| [13] |
Rosati C, Cadic A, Duron M, et al. Molecular characterization of the anthocyanidin synthase gene in Forsythia × intermedia reveals organ-specific expression during flower development[J]. Plant Sci, 1999, 149(1): 73-79.
doi: 10.1016/S0168-9452(99)00146-6 URL |
| [14] |
Ahn JH, Kim JS, Kim S, et al. De novo transcriptome analysis to identify anthocyanin biosynthesis genes responsible for tissue-specific pigmentation in zoysiagrass(Zoysia japonica steud.)[J]. PLoS One, 2015, 10(4): e0124497.
doi: 10.1371/journal.pone.0124497 URL |
| [15] |
Hong DY, Pan KY, Yu H. Taxonomy of the Paeonia delavayi complex(Paeoniaceae)[J]. Ann Mo Bot Gard, 1998, 85(4): 554.
doi: 10.2307/2992016 URL |
| [16] | 张艳丽, 李正红, 马宏, 等. 滇牡丹花色类群性状变异分析[J]. 植物分类与资源学报, 2011, 33(2): 183-190. |
| Zhang YL, Li ZH, Ma H, et al. A study on characters variation of different flower color groups of Paeonia delavayi(Paeoniac-eae)[J]. Plant Divers Resour, 2011, 33(2): 183-190. | |
| [17] | 张德全, 马阿丽, 杨永寿, 等. HPLC测定不同产地滇牡丹中没食子酸和丹皮酚含量[J]. 中国实验方剂学杂志, 2014, 20(5): 57-60. |
| Zhang DQ, Ma AL, Yang YS, et al. Content determination of trihydroxybenzoic acid and paeonol in Paeonia delavayi from different producing area by HPLC[J]. Chin J Exp Tradit Med Formulae, 2014, 20(5): 57-60. | |
| [18] |
Yoshida K, Mori M, Kondo T. Blue flower color development by anthocyanins: from chemical structure to cell physiology[J]. Nat Prod Rep, 2009, 26(7): 884-915.
doi: 10.1039/b800165k pmid: 19554240 |
| [19] |
Zhao DQ, Tao J. Recent advances on the development and regulation of flower color in ornamental plants[J]. Front Plant Sci, 2015, 6: 261.
doi: 10.3389/fpls.2015.00261 pmid: 25964787 |
| [20] |
Shi QQ, Zhou L, Wang Y, et al. Transcriptomic analysis of Paeonia delavayi wild population flowers to identify differentially expressed genes involved in purple-red and yellow petal pigmentation[J]. PLoS One, 2015, 10(8): e0135038.
doi: 10.1371/journal.pone.0135038 URL |
| [21] |
Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids[J]. Plant J, 2008, 54(4): 733-749.
doi: 10.1111/j.1365-313X.2008.03447.x URL |
| [22] | 李小兰, 张明生, 吕享. 植物花青素合成酶ANS基因的研究进展[J]. 植物生理学报, 2016, 52(6): 817-827. |
| Li XL, Zhang MS, Lü X. The research progress on plant anthocyanin synthetase ANS gene[J]. Plant Physiol J, 2016, 52(6): 817-827. | |
| [23] |
Liu Y, Shi Z, Maximova S, et al. Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase[J]. BMC Plant Biol, 2013, 13: 202.
doi: 10.1186/1471-2229-13-202 pmid: 24308601 |
| [24] | 陈跃华, 许云, 吴文嫱, 等. 参薯DaANS基因克隆及表达差异分析[J]. 植物生理学报, 2015, 51(6): 853-859. |
| Chen YH, Xu Y, Wu WQ, et al. Cloning and analysis of differential expression of DaANS gene in Dioscorea alata[J]. Plant Physiol J, 2015, 51(6): 853-859. | |
| [25] | 卜星星, 雒晓鹏, 白悦辰, 等. 金荞麦花青素合酶基因的克隆及其表达与花青素量的相关性研究[J]. 中草药, 2014, 45(7): 985-989. |
| Bu XX, Luo XP, Bai YC, et al. Gene cloning of anthocyanin synthase in Fagopyrum dibotrys and correlation between its expression level and anthocyanin content[J]. Chin Tradit Herb Drugs, 2014, 45(7): 985-989. | |
| [26] | 陈可欣, 王晓明, 曾慧杰, 等. 紫薇LiANS基因的克隆和表达及与花青素含量的关系[J]. 分子植物育种, 2021, 19(12): 3915-3922. |
| Chen KX, Wang XM, Zeng HJ, et al. Cloning and expression analysis of LiANS gene and its correlations with anthocyanin accumulation in Lagerstroemia indica[J]. Mol Plant Breed, 2021, 19(12): 3915-3922. | |
| [27] | 李果, 李厚华, 张延龙, 等. 凤丹牡丹ANS(PoANS)基因克隆、特性及表达[J]. 东北林业大学学报, 2016, 44(7): 64-69. |
| Li G, Li HH, Zhang YL, et al. Isolation, characterization and expression analysis of anthocyanidin synthase(ANS)gene in Paeonia ostii ‘Fengdan’[J]. J Northeast For Univ, 2016, 44(7): 64-69. | |
| [28] |
Collette VE, Jameson PE, Schwinn KE, et al. Temporal and spatial expression of flavonoid biosynthetic genes in flowers of Anthurium andraeanum[J]. Physiol Plant, 2004, 122(3): 297-304.
doi: 10.1111/ppl.2004.122.issue-3 URL |
| [29] | 张彬, 尹美强, 温银元, 等. 羽衣甘蓝花青素合成途径结构基因的表达特性[J]. 山西农业科学, 2014, 42(4): 313-316. |
| Zhang B, Yin MQ, Wen YY, et al. Expression features of the anthocyanin biosynthetic structural genes in kale[J]. J Shanxi Agric Sci, 2014, 42(4): 313-316. | |
| [30] |
Zhao DQ, Tao J, Han CX, et al. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony(Paeonia lactiflora Pall.)[J]. Mol Biol Rep, 2012, 39(12): 11263-11275.
doi: 10.1007/s11033-012-2036-7 URL |
| [31] | 华梅, 原晓龙, 杨卫, 等. HPLC分析6种不同花色滇牡丹花瓣中花青素和黄酮[J]. 西部林业科学, 2017, 46(6): 40-45. |
| Hua M, Yuan XL, Yang W, et al. Analysis of anthocyanins and flavonols in six different colors of petals of Paeonia delavayi by high performance liquid chromatography[J]. J West China For Sci, 2017, 46(6): 40-45. |
| [1] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [2] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [3] | 杨小峰, 秦小伟, 郭泽媛, 吕丽华. 原花青素对体外培养绵羊卵泡颗粒细胞增殖的影响[J]. 生物技术通报, 2022, 38(9): 258-263. |
| [4] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [5] | 于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193. |
| [6] | 陈佳敏, 刘永杰, 马锦绣, 李丹, 公杰, 赵昌平, 耿洪伟, 高世庆. 小麦组蛋白甲基化酶在杂交种中干旱胁迫表达模式分析[J]. 生物技术通报, 2022, 38(7): 51-61. |
| [7] | 段玥彤, 王鹏年, 张春宝, 林春晶. 植物黄烷酮-3-羟化酶基因研究进展[J]. 生物技术通报, 2022, 38(6): 27-33. |
| [8] | 王楠, 张瑞, 潘阳阳, 何翃宏, 王靖雷, 崔燕, 余四九. 牦牛TGF-β1基因克隆及在雌性生殖系统主要器官中的表达定位[J]. 生物技术通报, 2022, 38(6): 279-290. |
| [9] | 李宇航, 王兴平, 杨箭, 罗仍卓么, 任倩倩, 魏大为, 马云. miR-665在奶牛乳腺上皮细胞炎症中的表达及功能分析[J]. 生物技术通报, 2022, 38(5): 159-168. |
| [10] | 李洋, 张晓天, 朴静子, 周如军, 李自博, 关海雯. 花生疮痂病菌蓝光受体EaWC 1基因克隆及生物信息学分析[J]. 生物技术通报, 2022, 38(5): 93-99. |
| [11] | 邹良平, 郭鑫, 起登凤, 翟敏, 李壮, 赵平娟, 彭明, 牛兴奎. 低氮胁迫诱导木薯幼苗花青素积累及其基因表达[J]. 生物技术通报, 2022, 38(2): 75-82. |
| [12] | 张琳, 魏祯祯, 宋程威, 郭丽丽, 郭琪, 侯小改, 王华芳. ‘凤丹’牡丹PoFD基因克隆及表达分析[J]. 生物技术通报, 2022, 38(11): 104-111. |
| [13] | 赖恭梯, 阙秋霞, 潘若, 刘雨轩, 王琦, 赖谱富, 高慧颖, 赖呈纯. 刺葡萄查尔酮合成酶基因CHS对不同光质的响应及转录因子调控分析[J]. 生物技术通报, 2022, 38(11): 129-139. |
| [14] | 罗雅方, 朱春花, 肖钰婷, 李方全, 张江, 王玉书. 羽衣甘蓝类黄酮糖基转移酶基因的筛选及分析[J]. 生物技术通报, 2022, 38(11): 194-201. |
| [15] | 郑青波, 叶娜, 张哓兰, 包鹏甲, 王福彬, 任稳稳, 廖月姣, 阎萍, 潘和平. 天祝白牦牛退行期毛囊细胞亚群鉴定以及特征基因生物信息学分析[J]. 生物技术通报, 2022, 38(10): 262-272. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||