








生物技术通报 ›› 2023, Vol. 39 ›› Issue (6): 233-247.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1369
尹明华1,2,3,4( ), 余锾媛1, 肖心怡1, 王玉婷1
), 余锾媛1, 肖心怡1, 王玉婷1
收稿日期:2022-11-07
出版日期:2023-06-26
发布日期:2023-07-07
通讯作者:
尹明华同为本文通讯作者作者简介:尹明华,女,硕士,教授,研究方向:植物生物技术;E-mail: yinminghua04@163.com;
基金资助:
YIN Ming-hua1,2,3,4( ), YU Huan-yuan1, XIAO Xin-yi1, WANG Yu-ting1
), YU Huan-yuan1, XIAO Xin-yi1, WANG Yu-ting1
Received:2022-11-07
Published:2023-06-26
Online:2023-07-07
摘要:
为分析江西铅山红芽芋叶绿体基因组的结构组成情况,判定其在芋属中的进化位置及与同芋属叶绿体基因组的区别,为芋属物种鉴定、遗传多样性分析和资源保护提供相关依据。使用Illumina NovaSeq 6000测序平台对江西铅山红芽芋叶绿体基因组进行测序,通过生物信息学分析方法进行序列组装、注释和特征分析,并利用Geneious、MISA、MAFF、FASTTREE、CUSP、Chips和Codon W等生物信息学软件对其基因组结构和数目、密码子偏好性、序列重复、SSR 位点和系统发育进行分析。结果表明,江西铅山红芽芋叶绿体基因组大小为 16 2544 bp,呈现四分体结构。共包含 130个基因,其中84个编码蛋白质的基因,37个tRNA基因,8个核糖体rRNA基因,1个假基因。通过密码子偏好性分析,平均有效密码子数为45.60,ENC值小于45的基因39个,说明其密码子偏好性强。通过 SSR分析,检测到101个SSR 位点,其中单核苷酸重复最多(约占83.17%),且单核苷酸以A/T型为主。与近缘种比较,发现其叶绿体基因组序列高度保守,尤其蛋白质编码序列相似度极高。此外,系统发育分析发现江西铅山红芽芋与芋属、岩芋属聚为一支。本研究得到了江西铅山红芽芋的叶绿体基因组基本情况与系统发育位置,为江西铅山红芽芋的物种辨别、天然种群遗传多样性与功能基因组学提供前期研究铺垫。
尹明华, 余锾媛, 肖心怡, 王玉婷. 江西铅山红芽芋叶绿体基因组特征及系统发育分析[J]. 生物技术通报, 2023, 39(6): 233-247.
YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan[J]. Biotechnology Bulletin, 2023, 39(6): 233-247.
| 基因分类 Category | 基因分组 Gene group | 基因名称 Gene name |
|---|---|---|
| 光合作用 Photosynthesis | 光合系统I基因 Photosystem I | psaA、psaB、psaC、psaI、psaJ |
| 光合系统II基因 Photosystem II | psbA、psbB、psbC、psbD、psbE、psbF、psbH、psbI、psbK、psbL、psbM、psbT、psbZ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA、petB、petD、petG、petL、petN | |
| ATP合成酶基因 ATP synthase | atpA、atpB、atpE、atpF、atpH、atpI | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhA、ndhB(2)、ndhC、ndhD、ndhE、ndhF、ndhG、ndhH、ndhI、ndhJ、ndhK | |
| 二磷酸核酮糖羧化酶大亚基基因 Rubisco large subunit | rbcL | |
| 自我复制Self-replication | RNA聚合酶亚基基因RNA polymeras | rpoA、rpoB、rpoC1、rpoC2 |
| 核糖体小亚基基因 Ribosomal proteins(SSU) | rps11、rps12(2)、rps14、rps15、rps16、rps18、 rps19、rps2、rps3、rps4、rps7(2)、rps8 | |
| 核糖体大亚基基因 Ribosomal proteins(LSU) | rpl14、rpl16、rpl2(2)、rpl20、rpl22、rpl23(2)、rpl32、rpl33、rpl36、 | |
| 转运RNA基因 Transfer RNAs | trnC-GCA、trnD-GUC、trnF-GAA、trnG-GCC、trnH-GUG、trnI-CAU(2)、trnK-UUU、trnL-CAA(2)、trnL-UAA、trnL-UAG、trnM-CAU、trnN-GUU(2)、trnP-UGG、trnQ-UUG、trnR-ACG(2)、trnR-UCU、trnS-GCU、trnS-GGA、trnS-UGA、trnT-UGU、trnV-GAC(2)、trnV-UAC、trnW-CCA、trnY-GUA、trnfM-CAU | |
| 核糖体RNA基因 Ribosomal RNAs | rrn16(2)、rrn23(2)、rrn4.5(2)、rrn5(2) | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 蛋白酶Protease | clpP1 | |
| 乙酰辅酶 A 羧化酶Acetyl-CoA carboxylase | accD | |
| 翻译起始因子 Translation initiation factor | infA | |
| 包膜蛋白基因 Envelope membrane protein | cemA | |
| C型细胞色素合成基因 C-type cytochrome synthesis gene | ccsA | |
| 未知功能Unknown function | 假定叶绿体阅读框 Hypothetical chloroplast reading frames | ycf1、ycf2(2) |
表1 江西铅山红芽芋叶绿体基因组注释基因列表
Table 1 List of genes found in chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 基因分类 Category | 基因分组 Gene group | 基因名称 Gene name |
|---|---|---|
| 光合作用 Photosynthesis | 光合系统I基因 Photosystem I | psaA、psaB、psaC、psaI、psaJ |
| 光合系统II基因 Photosystem II | psbA、psbB、psbC、psbD、psbE、psbF、psbH、psbI、psbK、psbL、psbM、psbT、psbZ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA、petB、petD、petG、petL、petN | |
| ATP合成酶基因 ATP synthase | atpA、atpB、atpE、atpF、atpH、atpI | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhA、ndhB(2)、ndhC、ndhD、ndhE、ndhF、ndhG、ndhH、ndhI、ndhJ、ndhK | |
| 二磷酸核酮糖羧化酶大亚基基因 Rubisco large subunit | rbcL | |
| 自我复制Self-replication | RNA聚合酶亚基基因RNA polymeras | rpoA、rpoB、rpoC1、rpoC2 |
| 核糖体小亚基基因 Ribosomal proteins(SSU) | rps11、rps12(2)、rps14、rps15、rps16、rps18、 rps19、rps2、rps3、rps4、rps7(2)、rps8 | |
| 核糖体大亚基基因 Ribosomal proteins(LSU) | rpl14、rpl16、rpl2(2)、rpl20、rpl22、rpl23(2)、rpl32、rpl33、rpl36、 | |
| 转运RNA基因 Transfer RNAs | trnC-GCA、trnD-GUC、trnF-GAA、trnG-GCC、trnH-GUG、trnI-CAU(2)、trnK-UUU、trnL-CAA(2)、trnL-UAA、trnL-UAG、trnM-CAU、trnN-GUU(2)、trnP-UGG、trnQ-UUG、trnR-ACG(2)、trnR-UCU、trnS-GCU、trnS-GGA、trnS-UGA、trnT-UGU、trnV-GAC(2)、trnV-UAC、trnW-CCA、trnY-GUA、trnfM-CAU | |
| 核糖体RNA基因 Ribosomal RNAs | rrn16(2)、rrn23(2)、rrn4.5(2)、rrn5(2) | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 蛋白酶Protease | clpP1 | |
| 乙酰辅酶 A 羧化酶Acetyl-CoA carboxylase | accD | |
| 翻译起始因子 Translation initiation factor | infA | |
| 包膜蛋白基因 Envelope membrane protein | cemA | |
| C型细胞色素合成基因 C-type cytochrome synthesis gene | ccsA | |
| 未知功能Unknown function | 假定叶绿体阅读框 Hypothetical chloroplast reading frames | ycf1、ycf2(2) |
| 区域Area | 碱基长度 Base length/ bp | T/% | C/% | A/% | G/% | GC/% | AT偏斜值AT skew | GC 偏斜值GC skew |
|---|---|---|---|---|---|---|---|---|
| 大单拷贝区LSC | 89 814 | 33.58 | 17.59 | 32.04 | 16.79 | 34.38 | -0.023 5 | -0.023 3 |
| 小单拷贝区SSC | 22 074 | 36.02 | 15.12 | 34.97 | 13.89 | 29.01 | -0.014 8 | -0.042 4 |
| 反向重复序列a(IRa) | 25 328 | 29.01 | 21.95 | 28.59 | 20.44 | 42.40 | -0.007 3 | -0.035 6 |
| 反向重复序列b(IRb) | 25 328 | 28.59 | 20.44 | 29.01 | 21.95 | 42.40 | 0.007 3 | 0.035 6 |
| 总基因组 | 162 544 | 32.42 | 18.38 | 31.43 | 17.77 | 36.15 | -0.015 5 | -0.016 9 |
表2 江西铅山红芽芋叶绿体全基因组核苷酸组成
Table 2 Nucleotide composition of the whole chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 区域Area | 碱基长度 Base length/ bp | T/% | C/% | A/% | G/% | GC/% | AT偏斜值AT skew | GC 偏斜值GC skew |
|---|---|---|---|---|---|---|---|---|
| 大单拷贝区LSC | 89 814 | 33.58 | 17.59 | 32.04 | 16.79 | 34.38 | -0.023 5 | -0.023 3 |
| 小单拷贝区SSC | 22 074 | 36.02 | 15.12 | 34.97 | 13.89 | 29.01 | -0.014 8 | -0.042 4 |
| 反向重复序列a(IRa) | 25 328 | 29.01 | 21.95 | 28.59 | 20.44 | 42.40 | -0.007 3 | -0.035 6 |
| 反向重复序列b(IRb) | 25 328 | 28.59 | 20.44 | 29.01 | 21.95 | 42.40 | 0.007 3 | 0.035 6 |
| 总基因组 | 162 544 | 32.42 | 18.38 | 31.43 | 17.77 | 36.15 | -0.015 5 | -0.016 9 |
| 核苷酸类型 Nucleotide type | 简单重复序列 Simple sequence repeat | 重复序列个数 Number of SSR |
|---|---|---|
| 单核苷酸 Mononucleotide | A | 45 |
| C | 2 | |
| G | 1 | |
| T | 36 | |
| 二核苷酸 Dinucleotide | AT | 5 |
| TA | 10 | |
| 三核苷酸 Trinucleotide | TTA | 1 |
| 四核苷酸 Tetranucleotide | TATG | 1 |
表3 江西铅山红芽芋叶绿体基因组SSR 序列
Table 3 Simple sequence repeats(SSR)type of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 核苷酸类型 Nucleotide type | 简单重复序列 Simple sequence repeat | 重复序列个数 Number of SSR |
|---|---|---|
| 单核苷酸 Mononucleotide | A | 45 |
| C | 2 | |
| G | 1 | |
| T | 36 | |
| 二核苷酸 Dinucleotide | AT | 5 |
| TA | 10 | |
| 三核苷酸 Trinucleotide | TTA | 1 |
| 四核苷酸 Tetranucleotide | TATG | 1 |

图2 江西铅山红芽芋和11个近缘种叶绿体基因组的 IR 边界分析
Fig. 2 IR boundary analysis of the chloroplast genomes of C. cormosus cv. L. Schoot var. Cormosus ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species
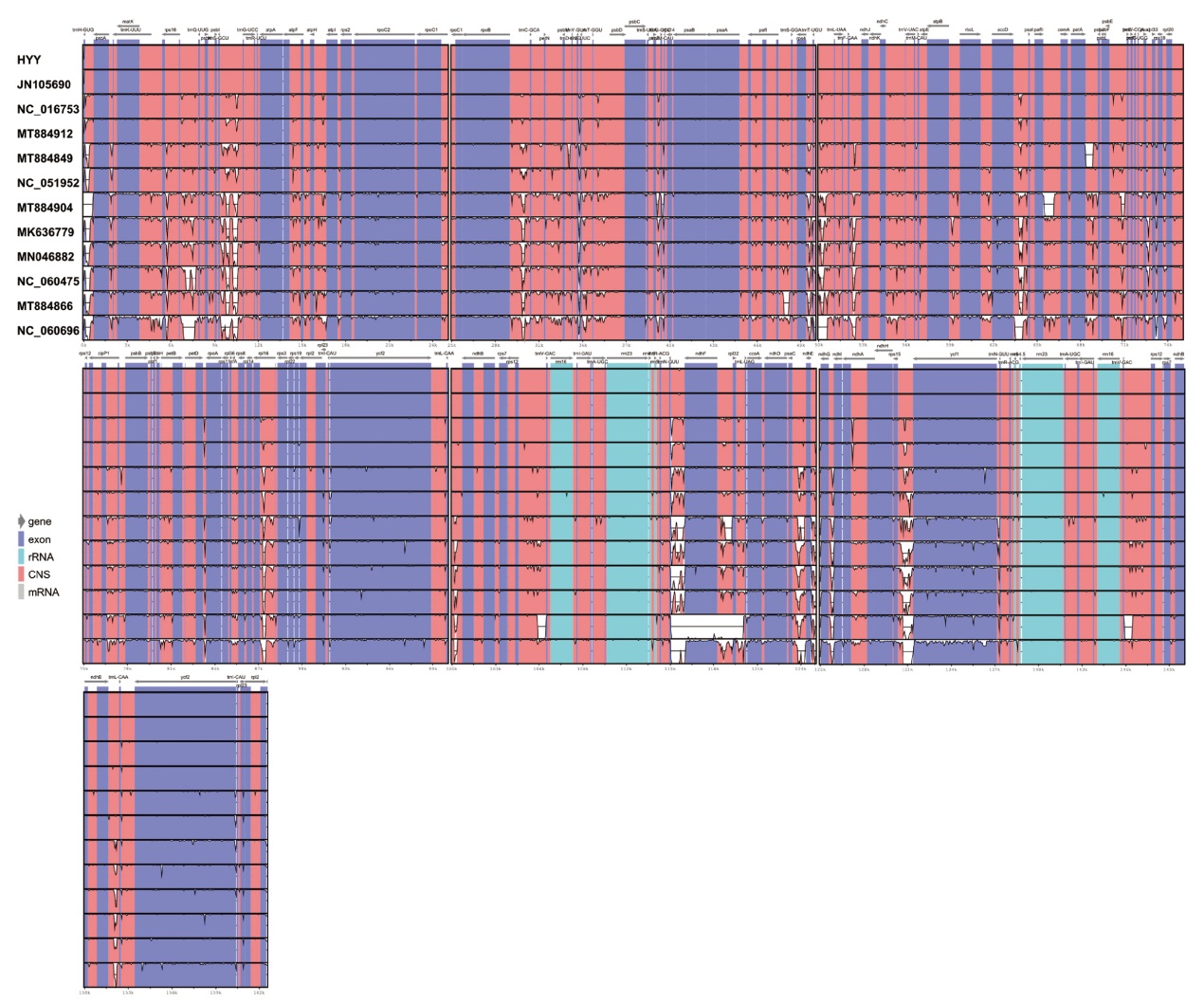
图3 江西铅山红芽芋和11种近缘种叶绿体基因组的可视化比对图
Fig. 3 Comparison of chloroplast genomes of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species

图4 江西铅山红芽芋和11种近缘种叶绿体基因组核苷酸多样性
Fig. 4 Nucleotide diversity of chloroplast genomes of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species
| 基因 Gene | GC含量 GC content/% | 基因 Gene | GC含量 GC content/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | ||||
| accD | 34.62 | 38.62 | 37.40 | 27.85 | 44.206 | psbH | 42.34 | 48.65 | 47.30 | 31.08 | 43.164 | ||
| atpA | 40.03 | 55.71 | 39.17 | 25.20 | 44.586 | psbI | 36.94 | 48.65 | 29.73 | 32.43 | 40.922 | ||
| atpB | 42.48 | 56.91 | 41.68 | 28.86 | 48.495 | psbJ | 41.46 | 41.46 | 53.66 | 29.27 | 20.769 | ||
| atpE | 41.67 | 52.94 | 41.91 | 30.15 | 46.888 | psbK | 37.57 | 42.86 | 36.51 | 33.33 | 42.160 | ||
| atpF | 37.73 | 50.00 | 35.16 | 28.02 | 39.808 | psbL | 29.06 | 35.90 | 28.21 | 23.08 | 50.514 | ||
| atpH | 44.31 | 62.20 | 48.78 | 21.95 | 49.183 | psbM | 28.57 | 42.86 | 25.71 | 17.14 | 31.500 | ||
| atpI | 38.04 | 49.19 | 37.90 | 27.02 | 43.152 | psbT | 35.29 | 41.18 | 35.29 | 29.41 | 34.772 | ||
| ccsA | 32.1 | 32.10 | 37.96 | 26.23 | 43.768 | psbZ | 34.39 | 38.10 | 42.86 | 22.22 | 51.110 | ||
| cemA | 31.45 | 39.62 | 27.04 | 27.67 | 40.639 | rbcL | 43.04 | 57.38 | 43.66 | 28.07 | 47.314 | ||
| clpP1 | 42.69 | 59.61 | 36.95 | 31.53 | 59.229 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| matK | 31.58 | 38.99 | 31.19 | 24.56 | 46.593 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| ndhA | 35.81 | 44.51 | 37.91 | 25.00 | 44.550 | rpl14 | 39.02 | 53.66 | 38.21 | 25.20 | 56.951 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl16 | 43.87 | 50.74 | 53.68 | 27.21 | 40.180 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl20 | 36.44 | 37.29 | 44.92 | 27.12 | 43.650 | ||
| ndhC | 37.47 | 49.59 | 33.88 | 28.93 | 45.634 | rpl22 | 37.63 | 45.16 | 40.32 | 27.42 | 51.065 | ||
| ndhD | 35.46 | 39.72 | 38.52 | 28.14 | 49.596 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhE | 31.37 | 38.24 | 33.33 | 22.55 | 42.469 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhF | 33.06 | 37.42 | 36.88 | 24.90 | 45.696 | rpl32 | 33.91 | 34.48 | 44.83 | 22.41 | 34.549 | ||
| ndhG | 33.15 | 42.37 | 33.33 | 23.73 | 43.846 | rpl33 | 34.33 | 38.81 | 38.81 | 25.37 | 42.754 | ||
| ndhH | 38.16 | 51.78 | 35.79 | 26.90 | 48.229 | rpl36 | 37.72 | 42.11 | 47.37 | 23.68 | 46.750 | ||
| ndhI | 34.44 | 43.89 | 36.67 | 22.78 | 44.213 | rpoA | 35.00 | 45.59 | 34.41 | 25.00 | 49.552 | ||
| ndhJ | 39.2 | 48.43 | 37.74 | 31.45 | 51.139 | rpoB | 39.05 | 50.23 | 37.74 | 29.17 | 49.169 | ||
| ndhK | 38.82 | 43.37 | 43.37 | 29.72 | 51.528 | rpoC1 | 38.40 | 50.29 | 38.16 | 26.75 | 49.009 | ||
| pafI | 39.45 | 47.34 | 40.83 | 30.18 | 55.939 | rpoC2 | 37.48 | 46.02 | 37.63 | 28.80 | 50.022 | ||
| pafII | 38.92 | 44.86 | 40.54 | 31.35 | 51.331 | rps2 | 38.96 | 43.88 | 43.04 | 29.96 | 45.542 | ||
| pbf1 | 46.21 | 52.27 | 43.18 | 43.18 | 48.475 | rps3 | 34.86 | 46.58 | 34.25 | 23.74 | 45.101 | ||
| petA | 40.5 | 53.58 | 35.51 | 32.4 | 51.521 | rps4 | 37.79 | 49.01 | 38.12 | 26.24 | 50.288 | ||
| petB | 39.66 | 47.22 | 41.67 | 30.09 | 42.013 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petD | 38.72 | 50.93 | 39.13 | 26.09 | 44.396 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petG | 38.6 | 55.26 | 31.58 | 28.95 | 33.100 | rps8 | 36.09 | 41.35 | 42.11 | 24.81 | 42.540 | ||
| petL | 37.5 | 37.50 | 46.88 | 28.12 | 35.182 | rps11 | 44.12 | 55.40 | 57.55 | 19.42 | 41.507 | ||
| petN | 41.11 | 50.00 | 43.33 | 30.00 | 30.698 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaA | 43.01 | 51.93 | 43.68 | 33.42 | 49.161 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaB | 40.91 | 48.44 | 42.99 | 31.29 | 48.022 | rps14 | 41.58 | 42.57 | 46.53 | 35.64 | 37.716 | ||
| psaC | 43.9 | 43.90 | 53.66 | 34.15 | 48.358 | rps15 | 34.05 | 40.86 | 29.03 | 32.26 | 49.599 | ||
| psaI | 35.14 | 43.24 | 27.03 | 35.14 | 44.365 | rps16 | 35.86 | 50.63 | 36.71 | 20.25 | 42.450 | ||
| psaJ | 35.66 | 41.86 | 39.53 | 25.58 | 42.629 | rps18 | 34.31 | 35.29 | 44.12 | 23.53 | 37.936 | ||
| psbA | 42.66 | 49.72 | 43.79 | 34.46 | 41.107 | rps19 | 35.84 | 43.01 | 39.78 | 24.73 | 42.905 | ||
| psbB | 43.42 | 54.42 | 46.37 | 29.47 | 48.296 | ycf1 | 30.85 | 37.27 | 30.03 | 25.25 | 47.451 | ||
| psbC | 44.37 | 54.33 | 46.54 | 32.25 | 45.499 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbD | 42.18 | 51.98 | 43.50 | 31.07 | 44.142 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbE | 40.08 | 42.86 | 46.43 | 30.95 | 48.400 | 平均 | 38.17 | 46.21 | 40.21 | 28.10 | 45.600 | ||
| psbF | 40.83 | 47.50 | 45.00 | 30.00 | 61.000 | Mean | |||||||
表4 江西铅山红芽芋叶绿体基因组48条CDS序列密码子的GC含量和有效密码子数
Table 4 GC content and ENC of 48 CDS codons from the chloroplast of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ in Jiangxi Yanshan
| 基因 Gene | GC含量 GC content/% | 基因 Gene | GC含量 GC content/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | ||||
| accD | 34.62 | 38.62 | 37.40 | 27.85 | 44.206 | psbH | 42.34 | 48.65 | 47.30 | 31.08 | 43.164 | ||
| atpA | 40.03 | 55.71 | 39.17 | 25.20 | 44.586 | psbI | 36.94 | 48.65 | 29.73 | 32.43 | 40.922 | ||
| atpB | 42.48 | 56.91 | 41.68 | 28.86 | 48.495 | psbJ | 41.46 | 41.46 | 53.66 | 29.27 | 20.769 | ||
| atpE | 41.67 | 52.94 | 41.91 | 30.15 | 46.888 | psbK | 37.57 | 42.86 | 36.51 | 33.33 | 42.160 | ||
| atpF | 37.73 | 50.00 | 35.16 | 28.02 | 39.808 | psbL | 29.06 | 35.90 | 28.21 | 23.08 | 50.514 | ||
| atpH | 44.31 | 62.20 | 48.78 | 21.95 | 49.183 | psbM | 28.57 | 42.86 | 25.71 | 17.14 | 31.500 | ||
| atpI | 38.04 | 49.19 | 37.90 | 27.02 | 43.152 | psbT | 35.29 | 41.18 | 35.29 | 29.41 | 34.772 | ||
| ccsA | 32.1 | 32.10 | 37.96 | 26.23 | 43.768 | psbZ | 34.39 | 38.10 | 42.86 | 22.22 | 51.110 | ||
| cemA | 31.45 | 39.62 | 27.04 | 27.67 | 40.639 | rbcL | 43.04 | 57.38 | 43.66 | 28.07 | 47.314 | ||
| clpP1 | 42.69 | 59.61 | 36.95 | 31.53 | 59.229 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| matK | 31.58 | 38.99 | 31.19 | 24.56 | 46.593 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| ndhA | 35.81 | 44.51 | 37.91 | 25.00 | 44.550 | rpl14 | 39.02 | 53.66 | 38.21 | 25.20 | 56.951 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl16 | 43.87 | 50.74 | 53.68 | 27.21 | 40.180 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl20 | 36.44 | 37.29 | 44.92 | 27.12 | 43.650 | ||
| ndhC | 37.47 | 49.59 | 33.88 | 28.93 | 45.634 | rpl22 | 37.63 | 45.16 | 40.32 | 27.42 | 51.065 | ||
| ndhD | 35.46 | 39.72 | 38.52 | 28.14 | 49.596 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhE | 31.37 | 38.24 | 33.33 | 22.55 | 42.469 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhF | 33.06 | 37.42 | 36.88 | 24.90 | 45.696 | rpl32 | 33.91 | 34.48 | 44.83 | 22.41 | 34.549 | ||
| ndhG | 33.15 | 42.37 | 33.33 | 23.73 | 43.846 | rpl33 | 34.33 | 38.81 | 38.81 | 25.37 | 42.754 | ||
| ndhH | 38.16 | 51.78 | 35.79 | 26.90 | 48.229 | rpl36 | 37.72 | 42.11 | 47.37 | 23.68 | 46.750 | ||
| ndhI | 34.44 | 43.89 | 36.67 | 22.78 | 44.213 | rpoA | 35.00 | 45.59 | 34.41 | 25.00 | 49.552 | ||
| ndhJ | 39.2 | 48.43 | 37.74 | 31.45 | 51.139 | rpoB | 39.05 | 50.23 | 37.74 | 29.17 | 49.169 | ||
| ndhK | 38.82 | 43.37 | 43.37 | 29.72 | 51.528 | rpoC1 | 38.40 | 50.29 | 38.16 | 26.75 | 49.009 | ||
| pafI | 39.45 | 47.34 | 40.83 | 30.18 | 55.939 | rpoC2 | 37.48 | 46.02 | 37.63 | 28.80 | 50.022 | ||
| pafII | 38.92 | 44.86 | 40.54 | 31.35 | 51.331 | rps2 | 38.96 | 43.88 | 43.04 | 29.96 | 45.542 | ||
| pbf1 | 46.21 | 52.27 | 43.18 | 43.18 | 48.475 | rps3 | 34.86 | 46.58 | 34.25 | 23.74 | 45.101 | ||
| petA | 40.5 | 53.58 | 35.51 | 32.4 | 51.521 | rps4 | 37.79 | 49.01 | 38.12 | 26.24 | 50.288 | ||
| petB | 39.66 | 47.22 | 41.67 | 30.09 | 42.013 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petD | 38.72 | 50.93 | 39.13 | 26.09 | 44.396 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petG | 38.6 | 55.26 | 31.58 | 28.95 | 33.100 | rps8 | 36.09 | 41.35 | 42.11 | 24.81 | 42.540 | ||
| petL | 37.5 | 37.50 | 46.88 | 28.12 | 35.182 | rps11 | 44.12 | 55.40 | 57.55 | 19.42 | 41.507 | ||
| petN | 41.11 | 50.00 | 43.33 | 30.00 | 30.698 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaA | 43.01 | 51.93 | 43.68 | 33.42 | 49.161 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaB | 40.91 | 48.44 | 42.99 | 31.29 | 48.022 | rps14 | 41.58 | 42.57 | 46.53 | 35.64 | 37.716 | ||
| psaC | 43.9 | 43.90 | 53.66 | 34.15 | 48.358 | rps15 | 34.05 | 40.86 | 29.03 | 32.26 | 49.599 | ||
| psaI | 35.14 | 43.24 | 27.03 | 35.14 | 44.365 | rps16 | 35.86 | 50.63 | 36.71 | 20.25 | 42.450 | ||
| psaJ | 35.66 | 41.86 | 39.53 | 25.58 | 42.629 | rps18 | 34.31 | 35.29 | 44.12 | 23.53 | 37.936 | ||
| psbA | 42.66 | 49.72 | 43.79 | 34.46 | 41.107 | rps19 | 35.84 | 43.01 | 39.78 | 24.73 | 42.905 | ||
| psbB | 43.42 | 54.42 | 46.37 | 29.47 | 48.296 | ycf1 | 30.85 | 37.27 | 30.03 | 25.25 | 47.451 | ||
| psbC | 44.37 | 54.33 | 46.54 | 32.25 | 45.499 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbD | 42.18 | 51.98 | 43.50 | 31.07 | 44.142 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbE | 40.08 | 42.86 | 46.43 | 30.95 | 48.400 | 平均 | 38.17 | 46.21 | 40.21 | 28.10 | 45.600 | ||
| psbF | 40.83 | 47.50 | 45.00 | 30.00 | 61.000 | Mean | |||||||
| 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | ||
|---|---|---|---|---|---|---|---|---|---|
| 丙氨酸 Ala | GCA | 420 | 1.187 28 | 赖氨酸 Lys | AAA | 1 036 | 1.48 318 | ||
| GCC | 215 | 0.607 774 | AAG | 361 | 0.516 822 | ||||
| GCG | 145 | 0.409 894 | 蛋氨酸Met | AUG | 596 | 1.993 31 | |||
| GCU | 635 | 1.795 05 | GUG | 2 | 0.006 688 96 | ||||
| 精氨酸 Arg | AGA | 521 | 1.953 75 | 苯丙氨酸Phe | UUC | 536 | 0.724 324 | ||
| AGG | 154 | 0.577 5 | UUU | 944 | 1.275 68 | ||||
| CGA | 358 | 1.342 5 | 脯氨酸 Pro | CCA | 322 | 1.194 81 | |||
| CGC | 96 | 0.36 | CCC | 197 | 0.730 983 | ||||
| CGG | 113 | 0.423 75 | CCG | 123 | 0.456 401 | ||||
| CGU | 358 | 1.342 5 | CCU | 436 | 1.617 81 | ||||
| 天冬酰胺 Asn | AAC | 290 | 0.464 | 丝氨酸 Ser | AGC | 104 | 0.298 851 | ||
| AAU | 960 | 1.536 | AGU | 447 | 1.284 48 | ||||
| 天冬氨酸 Asp | GAC | 213 | 0.394 81 | UCA | 444 | 1.275 86 | |||
| GAU | 866 | 1.605 19 | UCC | 339 | 0.974 138 | ||||
| 半胱氨酸 Cys | UGC | 74 | 0.485 246 | UCG | 179 | 0.514 368 | |||
| UGU | 231 | 1.514 75 | UCU | 575 | 1.652 3 | ||||
| 谷氨酰胺 Gln | CAA | 690 | 1.490 28 | 终止密码子Ter | UAA | 38 | 1.357 14 | ||
| CAG | 236 | 0.509 719 | UAG | 26 | 0.928 571 | ||||
| 谷氨酸 Glu | GAA | 1 035 | 1.483 87 | UGA | 20 | 0.714 286 | |||
| GAG | 360 | 0.516 129 | 苏氨酸 Thr | ACA | 433 | 1.277 29 | |||
| 甘氨酸 Gly | GGA | 719 | 1.632 24 | ACC | 247 | 0.728 614 | |||
| GGC | 161 | 0.365 494 | ACG | 149 | 0.439 528 | ||||
| GGG | 305 | 0.692 395 | ACU | 527 | 1.554 57 | ||||
| GGU | 577 | 1.309 88 | 色氨酸Trp | UGG | 443 | 1 | |||
| 组氨酸 His | CAC | 153 | 0.485 714 | 酪氨酸 Tyr | UAC | 208 | 0.428 424 | ||
| CAU | 477 | 1.514 29 | UAU | 763 | 1.571 58 | ||||
| 异亮氨酸 Ile | AUA | 702 | 0.950 79 | 缬氨酸 Val | GUA | 542 | 1.503 47 | ||
| AUC | 424 | 0.574 266 | GUC | 186 | 0.515 95 | ||||
| AUU | 1 089 | 1.474 94 | GUG | 194 | 0.538 141 | ||||
| 亮氨酸 Leu | CUA | 358 | 0.792 62 | GUU | 520 | 1.442 44 | |||
| CUC | 186 | 0.411 808 | |||||||
| CUG | 163 | 0.360 886 | |||||||
| CUU | 584 | 1.292 99 | |||||||
| UUA | 834 | 1.846 49 | |||||||
| UUG | 585 | 1.295 2 |
表5 江西铅山红芽芋叶绿体基因组中各氨基酸密码子的相对使用度
Table 5 RSCU analysis of amino acid codons in the chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | ||
|---|---|---|---|---|---|---|---|---|---|
| 丙氨酸 Ala | GCA | 420 | 1.187 28 | 赖氨酸 Lys | AAA | 1 036 | 1.48 318 | ||
| GCC | 215 | 0.607 774 | AAG | 361 | 0.516 822 | ||||
| GCG | 145 | 0.409 894 | 蛋氨酸Met | AUG | 596 | 1.993 31 | |||
| GCU | 635 | 1.795 05 | GUG | 2 | 0.006 688 96 | ||||
| 精氨酸 Arg | AGA | 521 | 1.953 75 | 苯丙氨酸Phe | UUC | 536 | 0.724 324 | ||
| AGG | 154 | 0.577 5 | UUU | 944 | 1.275 68 | ||||
| CGA | 358 | 1.342 5 | 脯氨酸 Pro | CCA | 322 | 1.194 81 | |||
| CGC | 96 | 0.36 | CCC | 197 | 0.730 983 | ||||
| CGG | 113 | 0.423 75 | CCG | 123 | 0.456 401 | ||||
| CGU | 358 | 1.342 5 | CCU | 436 | 1.617 81 | ||||
| 天冬酰胺 Asn | AAC | 290 | 0.464 | 丝氨酸 Ser | AGC | 104 | 0.298 851 | ||
| AAU | 960 | 1.536 | AGU | 447 | 1.284 48 | ||||
| 天冬氨酸 Asp | GAC | 213 | 0.394 81 | UCA | 444 | 1.275 86 | |||
| GAU | 866 | 1.605 19 | UCC | 339 | 0.974 138 | ||||
| 半胱氨酸 Cys | UGC | 74 | 0.485 246 | UCG | 179 | 0.514 368 | |||
| UGU | 231 | 1.514 75 | UCU | 575 | 1.652 3 | ||||
| 谷氨酰胺 Gln | CAA | 690 | 1.490 28 | 终止密码子Ter | UAA | 38 | 1.357 14 | ||
| CAG | 236 | 0.509 719 | UAG | 26 | 0.928 571 | ||||
| 谷氨酸 Glu | GAA | 1 035 | 1.483 87 | UGA | 20 | 0.714 286 | |||
| GAG | 360 | 0.516 129 | 苏氨酸 Thr | ACA | 433 | 1.277 29 | |||
| 甘氨酸 Gly | GGA | 719 | 1.632 24 | ACC | 247 | 0.728 614 | |||
| GGC | 161 | 0.365 494 | ACG | 149 | 0.439 528 | ||||
| GGG | 305 | 0.692 395 | ACU | 527 | 1.554 57 | ||||
| GGU | 577 | 1.309 88 | 色氨酸Trp | UGG | 443 | 1 | |||
| 组氨酸 His | CAC | 153 | 0.485 714 | 酪氨酸 Tyr | UAC | 208 | 0.428 424 | ||
| CAU | 477 | 1.514 29 | UAU | 763 | 1.571 58 | ||||
| 异亮氨酸 Ile | AUA | 702 | 0.950 79 | 缬氨酸 Val | GUA | 542 | 1.503 47 | ||
| AUC | 424 | 0.574 266 | GUC | 186 | 0.515 95 | ||||
| AUU | 1 089 | 1.474 94 | GUG | 194 | 0.538 141 | ||||
| 亮氨酸 Leu | CUA | 358 | 0.792 62 | GUU | 520 | 1.442 44 | |||
| CUC | 186 | 0.411 808 | |||||||
| CUG | 163 | 0.360 886 | |||||||
| CUU | 584 | 1.292 99 | |||||||
| UUA | 834 | 1.846 49 | |||||||
| UUG | 585 | 1.295 2 |
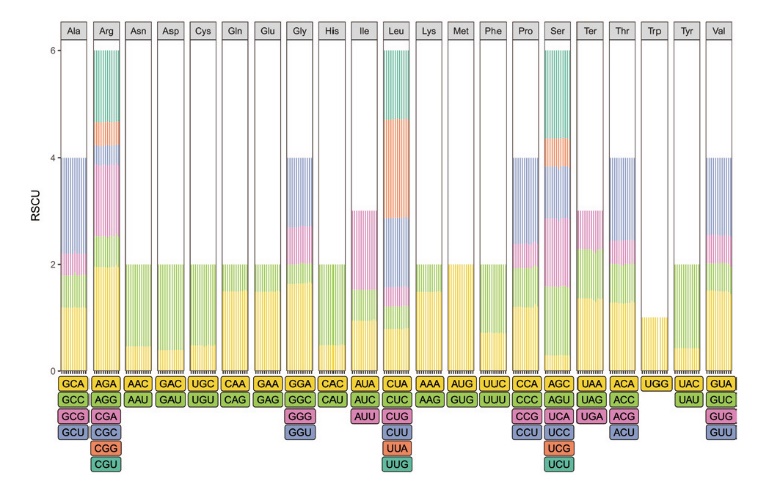
图6 江西铅山红芽芋叶绿体基因组密码子同义密码子相对使用度分布情况
Fig. 6 Distribution of codon RSCU in the chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
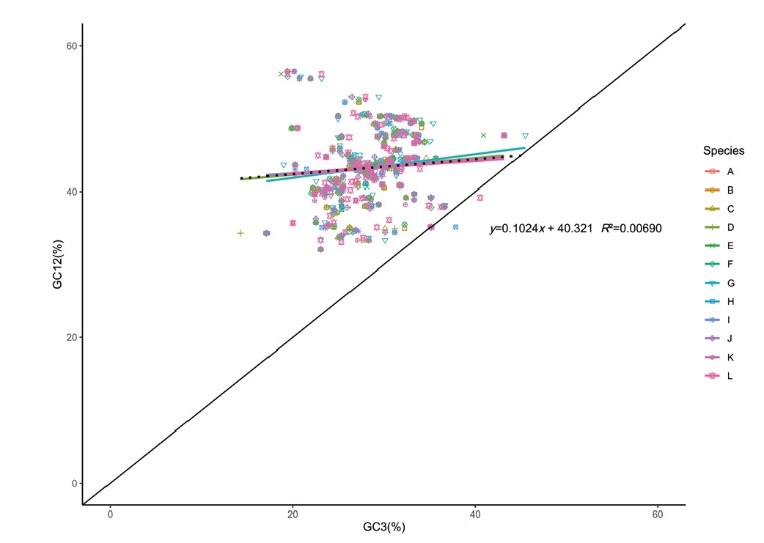
图7 江西铅山红芽芋和11种近缘种叶绿体基因中性绘图分析
Fig. 7 Neutrality plot analysis of chloroplast genes of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species A-L successively represents 11 related species(C. esculenta JN105690, C. esculenta NC_016753, R. vivipara MT884912, S. sp. 0020020418 MT884849, S. colocasiifolia NC_051952, A. peltata MT884904, A. fornicata MK636779, A. navicularis MN046882, L. gigantea NC_060475, E. hypnosum MT884866, A. cucullata NC_060696). The same below
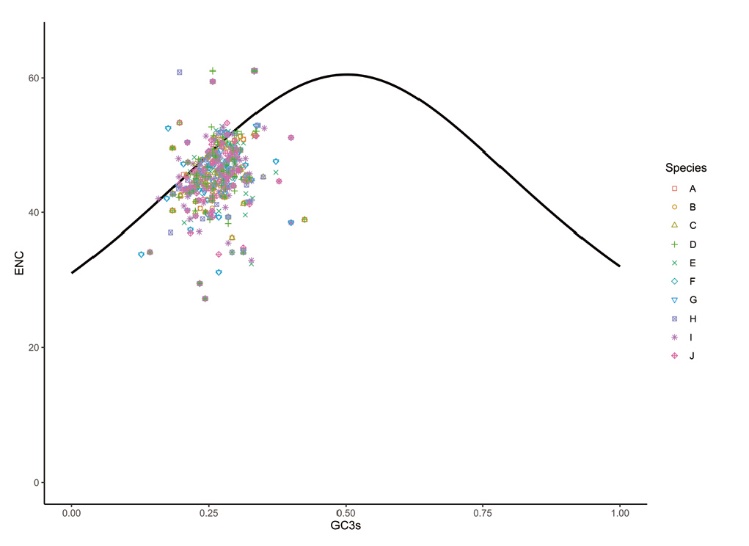
图8 江西铅山红芽芋和11种近缘种叶绿体基因组的ENC-plot分析
Fig. 8 ENC-plot analysis of chloroplast genes of C. esculenta L. Schoot var. Cormosus ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species
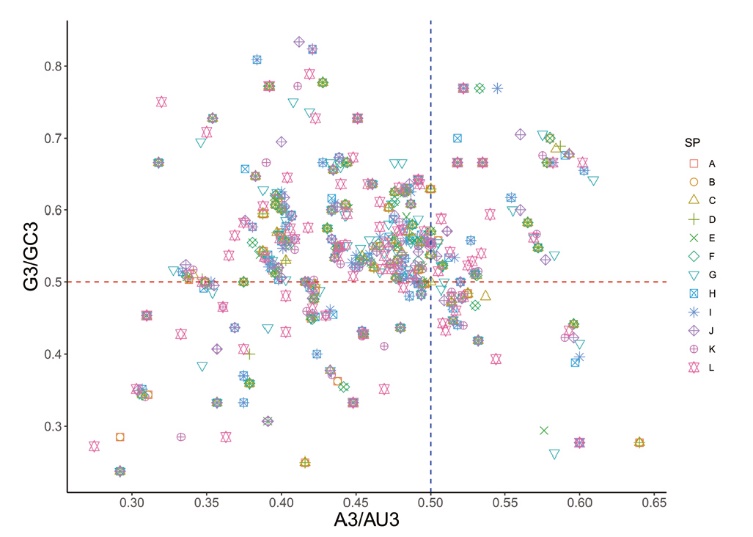
图9 江西铅山红芽芋和11种近缘种叶绿体基因组的PR2-plot 绘图分析
Fig. 9 PR2-plot analysis of chloroplast genes of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species
| 氨基酸 Amino acid | 密码子 Codon | 低表达基因 Low expressed gene | 高表达基因 High expressed gene | 相对使用度差值 △RSCU | |||
|---|---|---|---|---|---|---|---|
| 相对使用度 RSCU | 数量 Amount | 相对使用度 RSCU | 数量 Amount | ||||
| Ala | GCU | 1.854 3 | 70 | 2 | 8 | 0.145 7 | |
| Arg | CGU | 0.672 414 | 39 | 1.875 | 3 | 1.202 586 | |
| AGG | 0.879 31 | 51 | 1.25 | 2 | 0.370 69 | ||
| CGC | 0.379 31 | 22 | 0.625 | 1 | 0.245 69 | ||
| Asn | AAC | 0.520 376 | 83 | 1 | 3 | 0.479 624 | |
| Gln | CAA | 1.299 49 | 128 | 1.5 | 3 | 0.200 51 | |
| Gly | GGU | 0.967 442 | 52 | 1.312 5 | 7 | 0.345 058 | |
| Ile | AUU | 1.334 93 | 186 | 2 | 12 | 0.665 07 | |
| Leu | UUA | 1.043 48 | 92 | 1.555 56 | 7 | 0.512 08 | |
| CUU | 1.417 77 | 125 | 1.555 56 | 7 | 0.137 79 | ||
| CUA | 0.793 951 | 70 | 1.111 11 | 5 | 0.317 159 | ||
| Lys | AAA | 1.254 78 | 197 | 1.538 46 | 10 | 0.283 68 | |
| Phe | UUU | 1.032 84 | 173 | 1.666 67 | 10 | 0.633 83 | |
| Pro | CCU | 1.301 44 | 68 | 1.6 | 4 | 0.298 56 | |
| Ser | AGU | 0.928 433 | 80 | 1.263 16 | 6 | 0.334 727 | |
| UCA | 1.334 62 | 115 | 1.473 68 | 7 | 0.139 06 | ||
| Ter | UGA | 0 | 0 | 0.6 | 1 | 0.6 | |
| UAG | 0.666 667 | 2 | 1.2 | 2 | 0.533 333 | ||
| Thr | ACU | 1.192 66 | 65 | 1.384 62 | 9 | 0.191 96 | |
| Tyr | UAC | 0.402 174 | 37 | 0.666 667 | 2 | 0.264 493 | |
| Val | GUA | 1.106 8 | 57 | 2.117 65 | 9 | 1.010 85 | |
表6 江西铅山红芽芋叶绿体基因组最优密码子分析
Table 6 Putative optimal codons in chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 氨基酸 Amino acid | 密码子 Codon | 低表达基因 Low expressed gene | 高表达基因 High expressed gene | 相对使用度差值 △RSCU | |||
|---|---|---|---|---|---|---|---|
| 相对使用度 RSCU | 数量 Amount | 相对使用度 RSCU | 数量 Amount | ||||
| Ala | GCU | 1.854 3 | 70 | 2 | 8 | 0.145 7 | |
| Arg | CGU | 0.672 414 | 39 | 1.875 | 3 | 1.202 586 | |
| AGG | 0.879 31 | 51 | 1.25 | 2 | 0.370 69 | ||
| CGC | 0.379 31 | 22 | 0.625 | 1 | 0.245 69 | ||
| Asn | AAC | 0.520 376 | 83 | 1 | 3 | 0.479 624 | |
| Gln | CAA | 1.299 49 | 128 | 1.5 | 3 | 0.200 51 | |
| Gly | GGU | 0.967 442 | 52 | 1.312 5 | 7 | 0.345 058 | |
| Ile | AUU | 1.334 93 | 186 | 2 | 12 | 0.665 07 | |
| Leu | UUA | 1.043 48 | 92 | 1.555 56 | 7 | 0.512 08 | |
| CUU | 1.417 77 | 125 | 1.555 56 | 7 | 0.137 79 | ||
| CUA | 0.793 951 | 70 | 1.111 11 | 5 | 0.317 159 | ||
| Lys | AAA | 1.254 78 | 197 | 1.538 46 | 10 | 0.283 68 | |
| Phe | UUU | 1.032 84 | 173 | 1.666 67 | 10 | 0.633 83 | |
| Pro | CCU | 1.301 44 | 68 | 1.6 | 4 | 0.298 56 | |
| Ser | AGU | 0.928 433 | 80 | 1.263 16 | 6 | 0.334 727 | |
| UCA | 1.334 62 | 115 | 1.473 68 | 7 | 0.139 06 | ||
| Ter | UGA | 0 | 0 | 0.6 | 1 | 0.6 | |
| UAG | 0.666 667 | 2 | 1.2 | 2 | 0.533 333 | ||
| Thr | ACU | 1.192 66 | 65 | 1.384 62 | 9 | 0.191 96 | |
| Tyr | UAC | 0.402 174 | 37 | 0.666 667 | 2 | 0.264 493 | |
| Val | GUA | 1.106 8 | 57 | 2.117 65 | 9 | 1.010 85 | |
| [1] | 刘星月, 朱强龙, 李慧英, 等. 红芽芋脱毒试管芋诱导及植株再生[J]. 园艺学报, 2020, 47(12): 2427-2438. |
|
Liu XY, Zhu QL, Li HY, et al. Induction and plant regeneration of virus-free microtuber in red bud taro[J]. Acta Hortic Sin, 2020, 47(12): 2427-2438.
doi: 10.16420/j.issn.0513-353x.2020-0470 |
|
| [2] | 李云, 牛丽亚, 涂瑾, 等. 亲水胶体对红芽芋全粉理化特性和消化特性的影响[J]. 中国粮油学报, 2020, 35(2): 12-17. |
| Li Y, Niu LY, Tu J, et al. Effect of hydrocolloids on physicochemical properties and digestive properties of red bud taro flour[J]. J Chin Cereals Oils Assoc, 2020, 35(2): 12-17. | |
| [3] | 邓接楼, 曹昊玮, 李玲, 等. 脱毒红芽芋试管芋盆栽种植的农艺性状分析[J]. 分子植物育种, 2019, 17(22): 7500-7506. |
| Deng JL, Cao HW, Li L, et al. Analysis on agronomic traits of virus-free test-tube taro planted in pots[J]. Mol Plant Breed, 2019, 17(22): 7500-7506. | |
| [4] | 肖慈生, 方加军, 肖月土, 等. 铅山红芽芋早熟栽培技术规程[J]. 长江蔬菜, 2018(15): 29-30. |
| Xiao CS, Fang JJ, Xiao YT, et al. Technical regulations for early maturing cultivation of Yanshan red bud taro[J]. J Chang Veg, 2018(15): 29-30. | |
| [5] | 余志平, 林海红, 余俊红, 等. 铅山红芽芋产业发展概况[J]. 长江蔬菜, 2016(20): 34-36. |
| Yu ZP, Lin HH, Yu JH, et al. Development of red bud taro industry in Yanshan[J]. J Chang Veg, 2016(20): 34-36. | |
| [6] |
姜绍通, 程元珍, 郑志, 等. 红芽芋营养成分分析及评价[J]. 食品科学, 2012, 33(11): 269-272.
doi: 10.7506/spkx1002-6630-201211057 |
| Jiang ST, Cheng YZ, Zheng Z, et al. Analysis and evaluation of nutritional components of red bud taro(Colocasia esulenla L. schott)[J]. Food Sci, 2012, 33(11): 269-272. | |
| [7] |
Wang QR, Huang ZR, Gao CS, et al. The complete chloroplast genome sequence of Rubus hirsutus Thunb. and a comparative analysis within Rubus species[J]. Genetica, 2021, 149(5/6): 299-311.
doi: 10.1007/s10709-021-00131-9 |
| [8] |
Zhao ZY, Wang X, Yu Y, et al. Complete chloroplast genome sequences of Dioscorea: Characterization, genomic resources, and phylogenetic analyses[J]. PeerJ, 2018, 6: e6032.
doi: 10.7717/peerj.6032 URL |
| [9] |
Daniell H, Lin CS, Yu M, et al. Chloroplast genomes: diversity, evolution, and applications in genetic engineering[J]. Genome Biol, 2016, 17(1): 134.
doi: 10.1186/s13059-016-1004-2 pmid: 27339192 |
| [10] |
Ahmed I, Matthews PJ, Biggs PJ, et al. Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta(L.)Schott(Araceae)and closely related taxa[J]. Mol Ecol Resour, 2013, 13(5): 929-937.
doi: 10.1111/men.2013.13.issue-5 URL |
| [11] | Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data[J]. Nucleic Acids Res, 2017, 45(4): e18. |
| [12] |
Tillich M, Lehwark P, Pellizzer T, et al. GeSeq - versatile and accurate annotation of organelle genomes[J]. Nucleic Acids Res, 2017, 45(W1): W6-W11.
doi: 10.1093/nar/gkx391 URL |
| [13] |
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence[J]. Nucleic Acids Res, 1997, 25(5): 955-964.
doi: 10.1093/nar/25.5.955 pmid: 9023104 |
| [14] |
Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW(OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes[J]. Curr Genet, 2007, 52(5/6): 267-274.
doi: 10.1007/s00294-007-0161-y URL |
| [15] |
Thiel T, Michalek W, Varshney RK, et al. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley(Hordeum vulgare L.)[J]. Theor Appl Genet, 2003, 106(3): 411-422.
doi: 10.1007/s00122-002-1031-0 pmid: 12589540 |
| [16] |
Low SL, Yu CC, Ooi IH, et al. Extensive Miocene speciation in and out of Indochina: the biogeographic history of Typhonium sensu stricto(Araceae)and its implication for the assembly of Indochina flora[J]. J Syst Evol, 2021, 59(3): 419-428.
doi: 10.1111/jse.v59.3 URL |
| [17] |
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability[J]. Mol Biol Evol, 2013, 30(4): 772-780.
doi: 10.1093/molbev/mst010 pmid: 23329690 |
| [18] |
Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments[J]. PLoS One, 2010, 5(3): e9490.
doi: 10.1371/journal.pone.0009490 URL |
| [19] |
Sun LY, Jiang Z, Wan XX, et al. The complete chloroplast genome of Magnolia polytepala: comparative analyses offer implication for genetics and phylogeny of Yulania[J]. Gene, 2020, 736: 144410.
doi: 10.1016/j.gene.2020.144410 URL |
| [20] |
Ahmed I, Biggs PJ, Matthews PJ, et al. Mutational dynamics of aroid chloroplast genomes[J]. Genome Biol Evol, 2012, 4(12): 1316-1323.
doi: 10.1093/gbe/evs110 pmid: 23204304 |
| [21] |
Gao LQ, Li YL, Guo CC, et al. Indocalamus chongzhouensis(Poaceae: Bambusoideae), a new synonym of I. emeiensis: evidence from morphology and complete chloroplast genome data[J]. Phytotaxa, 2022, 542(1): 53-63.
doi: 10.11646/phytotaxa.542.1 URL |
| [22] |
Romero H, Zavala A, Musto H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces[J]. Nucleic Acids Res, 2000, 28(10): 2084-2090.
doi: 10.1093/nar/28.10.2084 pmid: 10773076 |
| [23] |
Carlini DB, Chen Y, Stephan W. The relationship between third-codon position nucleotide content, codon bias, mRNA secondary structure and gene expression in the drosophilid alcohol dehydrogenase genes Adh and Adhr[J]. Genetics, 2001, 159(2): 623-633.
doi: 10.1093/genetics/159.2.623 pmid: 11606539 |
| [24] |
Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis[J]. Proc Natl Acad Sci USA, 1999, 96(8): 4482-4487.
doi: 10.1073/pnas.96.8.4482 pmid: 10200288 |
| [25] |
Sharp PM, Emery LR, Zeng K. Forces that influence the evolution of codon bias[J]. Philos Trans R Soc Lond B Biol Sci, 2010, 365(1544): 1203-1212.
doi: 10.1098/rstb.2009.0305 URL |
| [26] |
Takahashi D, Sakaguchi S, Isagi Y, et al. Comparative chloroplast genomics of series Sakawanum in genus Asarum(Aristolochiaceae)to develop single nucleotide polymorphisms(SNPs)and simple sequence repeat(SSR)markers[J]. J For Res, 2018, 23(6): 387-392.
doi: 10.1080/13416979.2018.1518649 URL |
| [27] |
Yamane K, Kawahara T. Size homoplasy and mutational behavior of chloroplast simple sequence repeats(cpSSRs)inferred from intra- and interspecific variations in four chloroplast regions of diploid and polyploid Triticum and Aegilops species[J]. Genet Resour Crop Evol, 2018, 65(3): 727-743.
doi: 10.1007/s10722-017-0567-4 URL |
| [28] |
Dugas DV, Hernandez D, Koenen EJM, et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP[J]. Sci Rep, 2015, 5: 16958.
doi: 10.1038/srep16958 pmid: 26592928 |
| [29] |
Park S, An B, Park S. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation[J]. Sci Rep, 2018, 8(1): 13568.
doi: 10.1038/s41598-018-31938-w |
| [30] |
Raubeson LA, Peery R, Chumley TW, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus[J]. BMC Genomics, 2007, 8: 174.
pmid: 17573971 |
| [31] | Li X, Li Y, Zang M, et al. Complete chloroplast genome sequence and phylogenetic analysis of Quercus acutissima[J]. Int J Mol Sci, 2018, 19(8): E2443. |
| [32] |
Gu C, Ma L, Wu Z, et al. Comparative analyses of chloroplast genomes from 22 Lythraceae species: inferences for phylogenetic relationships and genome evolution within Myrtales[J]. BMC Plant Biol, 2019, 19(1): 281.
doi: 10.1186/s12870-019-1870-3 pmid: 31242865 |
| [33] |
Dong WP, Xu C, Li CH, et al. ycf1, the most promising plastid DNA barcode of land plants[J]. Sci Rep, 2015, 5: 8348.
doi: 10.1038/srep08348 pmid: 25672218 |
| [34] |
Amar MH. ycf1-ndhF genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica[J]. J Genet Eng Biotechnol, 2020, 18(1): 42.
doi: 10.1186/s43141-020-00057-3 |
| [1] | 张广志, 王加宁, 吴晓青, 周方园, 张新建, 赵晓燕, 谢雪迎, 周红姿. 设施番茄根围土样中木霉菌多样性及功能活性分析[J]. 生物技术通报, 2018, 34(4): 179-185. |
| [2] | 蔡东梅,龚国利. 大肠杆菌中生物药物的生产现状及展望[J]. 生物技术通报, 2016, 32(8): 34-40. |
| [3] | 胡文哲, 谭泽文, 王勇, 徐羡微, 谭志远. 藤县药用野生稻内生固氮菌分离鉴定及系统发育分析[J]. 生物技术通报, 2016, 32(6): 111-119. |
| [4] | 刘国红, 刘波, 林营志, 唐建阳. 基于脂肪酸生物标记与16S rRNA的芽胞杆菌系统发育分析比较[J]. 生物技术通报, 2015, 31(3): 146-153. |
| [5] | 倪玉佳, 周旻昱, 欧阳嘉, 郑兆娟, 勇强. 黑曲霉嗜热β-甘露聚糖酶在毕赤酵母中的克隆表达及其魔芋降解产物分析[J]. 生物技术通报, 2014, 0(6): 181-186. |
| [6] | 王菲, 刘抗, 范家同, 成守亮, 王子元. 一株耐盐弧菌Vibrio sp.K1-L的分离与鉴定[J]. 生物技术通报, 2014, 0(4): 127-131. |
| [7] | 侯新东, 尹帅, 盛桂莲, 程丹丹, 赖旭龙. 基于ITS序列探讨10种荨麻科植物的系统发育关系[J]. 生物技术通报, 2013, 0(8): 68-73. |
| [8] | 彭慧超;程大志;王爱珍;王环;王海庆;沈建伟;司庆文;韩发;周党卫;. 冬虫夏草培养子实体ITS,5.8S的分析及系统发育研究[J]. , 2010, 0(07): 128-133. |
| [9] | 王松华;田亚平;. 产酸性脲酶菌株的筛选、鉴定及其脲酶的应用初探[J]. , 2008, 0(06): 175-178. |
| [10] | 郝云婕;韩素贞;. gyrB基因在细菌系统发育分析中的应用[J]. , 2008, 0(02): 39-41. |
| [11] | 刘妍;李志勇;. 具有多重酶活性的澳大利亚厚皮海绵共附生放线菌的研究[J]. , 2006, 0(05): 121-125. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||