








生物技术通报 ›› 2023, Vol. 39 ›› Issue (6): 217-232.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1111
郭怡婷1,2( ), 赵文菊1,2, 任延靖1,2,3, 赵孟良1,2,3(
), 赵文菊1,2, 任延靖1,2,3, 赵孟良1,2,3( )
)
收稿日期:2022-09-09
出版日期:2023-06-26
发布日期:2023-07-07
通讯作者:
赵孟良,男,博士,副研究员,研究方向:蔬菜遗传育种与分子生物学;E-mail: 8304269@163.com作者简介:郭怡婷,女,硕士研究生,研究方向:蔬菜遗传育种与分子生物学;E-mail: yiting0201@163.com
基金资助:
GUO Yi-ting1,2( ), ZHAO Wen-ju1,2, REN Yan-jing1,2,3, ZHAO Meng-liang1,2,3(
), ZHAO Wen-ju1,2, REN Yan-jing1,2,3, ZHAO Meng-liang1,2,3( )
)
Received:2022-09-09
Published:2023-06-26
Online:2023-07-07
摘要:
通过转录组学的方法,筛选与菊芋叶片响应干旱胁迫密切相关的NAC转录因子,为今后菊芋NAC基因的克隆及功能验证奠定理论基础。以抗旱青芋2号菊芋品种为实验材料,采取25% PEG6000渗透模拟干旱胁迫的方法,分别在处理0、18、24和36 h后取菊芋叶片,通过转录组测序的方法,筛选干旱胁迫下菊芋叶片中差异表达的NAC转录因子,并对其进行分析。在菊芋叶片中共鉴定出了70个NAC基因,序列长度在408-1 869 bp之间,检测到的motif基序的长度在21-50个碱基不等,系统进化树分析显示,大部分的HtNAC基因能够与其他的NAC基因聚为一类;通过与已报道的抗旱相关的其他作物中的NAC基因进行聚类发现,HtNAC90-3与AtNAC72聚为一类,HtNAC67-3与AtNAC055聚为一类,HtNAC2-1与AtNAC019聚为一类,HtNAC36与SINAC6聚为一类,HtNAC83-1与AtNAC96聚为一类;组织特异性表达分析结果显示,HtNACs基因在种子和花瓣中的表达量相对较高,干旱诱导表达分析显示,分别有38个和11个HtNACs基因在菊芋叶片和根中呈现出诱导后上升表达的趋势。本研究发现HtNACs在种子和花瓣中表达量较高,尤其以花瓣中的表达量最高,后续可从菊芋的花瓣中克隆获得菊芋HtNACs并进行相关功能验证等工作。
郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232.
GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L.[J]. Biotechnology Bulletin, 2023, 39(6): 217-232.
| 引物名称Primer name | 上游引物Forward primer sequence(5'-3') | 下游引物Reserved primer sequence(5'-3') |
|---|---|---|
| Cluster-10091.0 | CAAATGAAAACAACACTACTCAGGAA | TTCCTGAGTAGTGTTGTTTTCATTTG |
| Cluster-10760.100264 | CATTTCAGAGCTAACGAGGACG | TTTAACCCGTGACTATAGGACCC |
| Cluster-10760.101731 | AGTGGCAAGAAGTGAAGTGAGTAA | AAGTAGCATAGAATACGGTGGTTAG |
| Cluster-10760.101956 | CTCCAAACAATCTCCGGTCAT | ATAACCCTCTTCATCCTTCTTCAC |
| Cluster-10760.106687 | TCGGTGAGCAGGAATGGTTCT | TCGGTCTTTTCGCCTTTAGGA |
| Cluster-10760.64992 | ATTCGTTTTCACGCCTTTTG | GGAGTGGTACTTCTTTTCGCC |
| Cluster-10760.63798 | GTGATGACGTGTGGCGGTT | TACTCTGAGACGCACATTTCCG |
| Cluster-10760.110871 | GGGTGATTAGCAGGGTTTTTC | GTTCTTCCATCGATTCGGAG |
| Cluster-10760.114800 | CTCCAACTGTCGTTTACTCATCA | TTTCCATTTAGTCTTCTTCCCTG |
| Cluster-10760.117836 | TGGGTTTTATGTCGTCTTTACAAC | TTCCCATCATCAATATCCTCTTTT |
| Cluster-10760.117837 | TTTCTAAGTTGGATGATTGGGTT | GTAGGATGAAGATGATGAATGTTGT |
| Cluster-10760.121177 | ATACGCGATATTTCTTTCACAGG | ATTACCCTCACTTTGCTCCAAC |
| Cluster-10760.125324 | TTATTACGCATTATCTCACGCC | TTCCCCCATCTTTGCTCTCT |
| Cluster-10760.125325 | CCCTATTTCTTCCCTTCATTTTC | CACCTTCGTTCACCACCGT |
| Cluster-10760.12609 | TAGAGTCCTGTCGGTTCTGTTG | ACATGTTCCTATCTTTTTCTGGTTA |
| Cluster-10760.126290 | CTGCCCTAAATCAAAAAAGGAT | AAGCACCAACTGGACAAACC |
| Cluster-10760.129386 | GTTCTTCCCTTTCTTGTCTTGG | AACTCATTTTGTTCTACCTCTCGA |
| Cluster-10760.130708 | CAACACACCAAGAACAGAAAAAT | GTCCCGAAAATATGGAAAATAAT |
| Cluster-10760.130718 | TGGAAAGCCACAGGAAAAGAC | AATCAGGCGAGACCCAAAAC |
| Cluster-10760.36796 | TGAAAGATTACAAAAGGCTGAGA | GAGTTATGGTGCTTGAAAGGC |
| Cluster-10760.133271 | TCACCAACAAAATCAGTAACAAAGA | CCGACAAAGGTAGTGAACGATG |
| Cluster-10760.134161 | CAGGCTCGCACTCTCTTATGA | TGGTTCGATCTGTGTTTTGTTG |
| Cluster-10760.135520 | GGCAAATAATGAAATGAGCAGG | TGGCGAGACAAGAGTGGATACA |
| Cluster-10760.135805 | CAACAACCTCAATCTAAACCAAA | GTTCAAGACCCATATCTACGACC |
| Cluster-10760.137050 | TCACACCAAGAGACCGGAAATA | ATCGTCGAGCCTGGAAACTG |
| Cluster-10760.138345 | GGAAGGACCGAAAGGTGAAC | CATCAAGACGATACTCATGCATAA |
| Cluster-10760.16091 | ATTCGACCATATTCGAGCGTT | TCTGATCCTTGGGAGTTACCAG |
| Cluster-10760.91122 | GCGTGGGAGGCAAGGAAT | TCAAGTTGAGGTGATTGAGGATCT |
| Cluster-10760.17171 | GGGTGAGTGGGTGATAAGTAAGG | GTCGGTCAAGAGAATCGAGGA |
| Cluster-10760.19944 | CGTTTGCTTTGTGTCGTGTGA | GTGCCCTTCTTGTCCTTTTGAT |
| Cluster-10760.26307 | CAAAAAGATGGTTTTAGGGAAGT | ATAGTGGACAAGAATCAGGCAGT |
| Cluster-10760.2853 | ACACAATCCCTCTCACCTTCC | ACCATCTCTGTTGACCCTATCC |
| Cluster-10760.34552 | CCACAAAGAAAGAAACAACCAA | CGCCTCAAGTAATACCTCACAA |
| Cluster-10760.40189 | CTGGTTAGCAGAGTATTGGTGG | CTTTGACTTCGTATATGAATTGGA |
| Cluster-10760.41713 | AAGCAAGAGTTGCAGATGAGAAG | CACTAGAGCCACAAGACGATGA |
| Cluster-10760.4894 | TGAAGGCTAAGGTTGGATGTGA | CGAAGGACGGTGAAAGAAGTG |
| Cluster-10760.49107 | CCGTACACCATAAAGGCCAAC | TCCCTCCAATTCCTGATCAGA |
| Cluster-10760.53392 | CGGTTTCAGGTTTCACCCAA | CGGCACATTTTTTTTCTCGTC |
| Cluster-10760.66624 | ATCTTTTGTCTCCCTCCGTTT | GACCCATGTTGGTTGATTTTG |
| Cluster-10760.54233 | AAGTAGAAGTGGAAGTGGGAGTTG | CATCAAGGACAACTGAGGTGTG |
| Cluster-10760.54351 | GATCTTCATCATGTTATCCCCGT | GCTTCCCTCCTAGAACTTCCTTT |
| Cluster-10760.54670 | GCAACATAAGTTTGGTAGAGGC | GAAGAGGTGAACCCGAGAGA |
| Cluster-10760.57561 | TAAACAAGAAGTGGAGGGGAC | GCCAATAAATCAACATGAACAAA |
| Cluster-10760.60147 | TCCTTACCAACATAAACCCTCA | TAGAAAATATCTCACAAGCCGC |
| Cluster-10760.63247 | TTTGGAATGTTTGGATGTTGAT | ATAGATTGTAGCTTACGGGGAGT |
| Cluster-10760.63290 | GTCGGTTTACCATCGGTCTCAA | GCTCATCATCCTCCCATTCCTC |
| Cluster-10760.65303 | TTTACAAAGGAAAACCCCCAC | CCACTTGAACCCGAAGATGA |
| Cluster-10760.70530 | CGACGACGAGAAAGAAAGAGAG | AGCCCGAAGATGATGAAACC |
| Cluster-10760.67533 | GGGACGCAAGGTCGGAAT | GTCGAGTTCGGCGGGTTAG |
| Cluster-10760.67861 | GAAGGAAGGAACCCATCAAGT | CAAAATCCAGAGAAACGCAGA |
| Cluster-10760.6999 | CAGGAAGAAGGATGGGTAGTGT | TCGAGCTGTGGGAGTTGTATAA |
| Cluster-10760.70147 | TGTTGGTCTAGTTGGTGTTTCC | TTTTTGTTCGTAGTTCAGTGTGC |
| Cluster-10760.73033 | AAAAGATGAGAACGGCGAGAGT | AGGATGCGTAATAGGGGAAAAC |
| Cluster-10760.73468 | GATTCTTGGTTTTCTTCTGCTTT | CTACTTGATCTCGACGGTTTTG |
| Cluster-10760.73971 | ACTTTTGTGTTTTATTTGGGGG | TGAGGTGTTGTATTGGTGGTGA |
| Cluster-10760.79303 | ATCAAGCGGTAGTGGTGGC | GGTTTGGGGTAGGAAATGTGT |
| Cluster-10760.81770 | CTCTCTCTTACCATACCACCCC | GATTTCCAATACCCGCTACTC |
| Cluster-10760.89696 | TGTGTAAATAGGGGAGTGGGAG | AGGTGTCGGTTGGTAAAGTAGG |
| Cluster-10760.9534 | CCCAAATCCTATCAACTCCATC | GAATCCTTGCTTCCTTCACTCA |
| Cluster-1169.0 | CATTTTCCTCACATAGGCCAC | CACCCACCGATACATCATCAC |
| Cluster-13332.0 | TCGTCTAAGGGTTGTTGTTCAC | AGCAATTTCTCATTCTTGGGTAT |
| Cluster-17703.0 | GTTCTCGCACAAATAGAGCTACC | AAAGTCTTTCTCATTCCAACCATA |
| Cluster-22290.1 | ACACCAACGGCAACAATAGAAA | CAAGAGTAGAGGCATGGAACGA |
| Cluster-23414.0 | GGAGAGCGTTGTCAAGGAGAC | GGAACTAGTGAGGCGGGATG |
| Cluster-24277.0 | GGTGTTGGAGCAGAGGGC | GCATAGGGGGGAGGAGTGA |
| Cluster-2502.0 | GATAAGGAGATCTTAAGGGGGC | GAGAAGCAGTGTACGTGAGTCG |
| Cluster-26181.0 | TTTTGTAAGCCGAGTTGTGGTA | TCAGATCGTCATCGAAAGTGAT |
| Cluster-27488.0 | CCCCAAGGGTTTTTTATGTGT | ATGTCGGACTGTGGATTTTTTC |
| Cluster-6808.0 | ACGTCGGTAAAGCTCCAAAAG | GGTGCTAGGACCACCAAAAAT |
| Cluster-7497.0 | TAAGTTTATGCCGGGTGTATGTG | GTTGTTGTGAAAGGGTTGTGGA |
表1 实时荧光定量PCR引物
Table 1 Real-time quantitative PCR primer
| 引物名称Primer name | 上游引物Forward primer sequence(5'-3') | 下游引物Reserved primer sequence(5'-3') |
|---|---|---|
| Cluster-10091.0 | CAAATGAAAACAACACTACTCAGGAA | TTCCTGAGTAGTGTTGTTTTCATTTG |
| Cluster-10760.100264 | CATTTCAGAGCTAACGAGGACG | TTTAACCCGTGACTATAGGACCC |
| Cluster-10760.101731 | AGTGGCAAGAAGTGAAGTGAGTAA | AAGTAGCATAGAATACGGTGGTTAG |
| Cluster-10760.101956 | CTCCAAACAATCTCCGGTCAT | ATAACCCTCTTCATCCTTCTTCAC |
| Cluster-10760.106687 | TCGGTGAGCAGGAATGGTTCT | TCGGTCTTTTCGCCTTTAGGA |
| Cluster-10760.64992 | ATTCGTTTTCACGCCTTTTG | GGAGTGGTACTTCTTTTCGCC |
| Cluster-10760.63798 | GTGATGACGTGTGGCGGTT | TACTCTGAGACGCACATTTCCG |
| Cluster-10760.110871 | GGGTGATTAGCAGGGTTTTTC | GTTCTTCCATCGATTCGGAG |
| Cluster-10760.114800 | CTCCAACTGTCGTTTACTCATCA | TTTCCATTTAGTCTTCTTCCCTG |
| Cluster-10760.117836 | TGGGTTTTATGTCGTCTTTACAAC | TTCCCATCATCAATATCCTCTTTT |
| Cluster-10760.117837 | TTTCTAAGTTGGATGATTGGGTT | GTAGGATGAAGATGATGAATGTTGT |
| Cluster-10760.121177 | ATACGCGATATTTCTTTCACAGG | ATTACCCTCACTTTGCTCCAAC |
| Cluster-10760.125324 | TTATTACGCATTATCTCACGCC | TTCCCCCATCTTTGCTCTCT |
| Cluster-10760.125325 | CCCTATTTCTTCCCTTCATTTTC | CACCTTCGTTCACCACCGT |
| Cluster-10760.12609 | TAGAGTCCTGTCGGTTCTGTTG | ACATGTTCCTATCTTTTTCTGGTTA |
| Cluster-10760.126290 | CTGCCCTAAATCAAAAAAGGAT | AAGCACCAACTGGACAAACC |
| Cluster-10760.129386 | GTTCTTCCCTTTCTTGTCTTGG | AACTCATTTTGTTCTACCTCTCGA |
| Cluster-10760.130708 | CAACACACCAAGAACAGAAAAAT | GTCCCGAAAATATGGAAAATAAT |
| Cluster-10760.130718 | TGGAAAGCCACAGGAAAAGAC | AATCAGGCGAGACCCAAAAC |
| Cluster-10760.36796 | TGAAAGATTACAAAAGGCTGAGA | GAGTTATGGTGCTTGAAAGGC |
| Cluster-10760.133271 | TCACCAACAAAATCAGTAACAAAGA | CCGACAAAGGTAGTGAACGATG |
| Cluster-10760.134161 | CAGGCTCGCACTCTCTTATGA | TGGTTCGATCTGTGTTTTGTTG |
| Cluster-10760.135520 | GGCAAATAATGAAATGAGCAGG | TGGCGAGACAAGAGTGGATACA |
| Cluster-10760.135805 | CAACAACCTCAATCTAAACCAAA | GTTCAAGACCCATATCTACGACC |
| Cluster-10760.137050 | TCACACCAAGAGACCGGAAATA | ATCGTCGAGCCTGGAAACTG |
| Cluster-10760.138345 | GGAAGGACCGAAAGGTGAAC | CATCAAGACGATACTCATGCATAA |
| Cluster-10760.16091 | ATTCGACCATATTCGAGCGTT | TCTGATCCTTGGGAGTTACCAG |
| Cluster-10760.91122 | GCGTGGGAGGCAAGGAAT | TCAAGTTGAGGTGATTGAGGATCT |
| Cluster-10760.17171 | GGGTGAGTGGGTGATAAGTAAGG | GTCGGTCAAGAGAATCGAGGA |
| Cluster-10760.19944 | CGTTTGCTTTGTGTCGTGTGA | GTGCCCTTCTTGTCCTTTTGAT |
| Cluster-10760.26307 | CAAAAAGATGGTTTTAGGGAAGT | ATAGTGGACAAGAATCAGGCAGT |
| Cluster-10760.2853 | ACACAATCCCTCTCACCTTCC | ACCATCTCTGTTGACCCTATCC |
| Cluster-10760.34552 | CCACAAAGAAAGAAACAACCAA | CGCCTCAAGTAATACCTCACAA |
| Cluster-10760.40189 | CTGGTTAGCAGAGTATTGGTGG | CTTTGACTTCGTATATGAATTGGA |
| Cluster-10760.41713 | AAGCAAGAGTTGCAGATGAGAAG | CACTAGAGCCACAAGACGATGA |
| Cluster-10760.4894 | TGAAGGCTAAGGTTGGATGTGA | CGAAGGACGGTGAAAGAAGTG |
| Cluster-10760.49107 | CCGTACACCATAAAGGCCAAC | TCCCTCCAATTCCTGATCAGA |
| Cluster-10760.53392 | CGGTTTCAGGTTTCACCCAA | CGGCACATTTTTTTTCTCGTC |
| Cluster-10760.66624 | ATCTTTTGTCTCCCTCCGTTT | GACCCATGTTGGTTGATTTTG |
| Cluster-10760.54233 | AAGTAGAAGTGGAAGTGGGAGTTG | CATCAAGGACAACTGAGGTGTG |
| Cluster-10760.54351 | GATCTTCATCATGTTATCCCCGT | GCTTCCCTCCTAGAACTTCCTTT |
| Cluster-10760.54670 | GCAACATAAGTTTGGTAGAGGC | GAAGAGGTGAACCCGAGAGA |
| Cluster-10760.57561 | TAAACAAGAAGTGGAGGGGAC | GCCAATAAATCAACATGAACAAA |
| Cluster-10760.60147 | TCCTTACCAACATAAACCCTCA | TAGAAAATATCTCACAAGCCGC |
| Cluster-10760.63247 | TTTGGAATGTTTGGATGTTGAT | ATAGATTGTAGCTTACGGGGAGT |
| Cluster-10760.63290 | GTCGGTTTACCATCGGTCTCAA | GCTCATCATCCTCCCATTCCTC |
| Cluster-10760.65303 | TTTACAAAGGAAAACCCCCAC | CCACTTGAACCCGAAGATGA |
| Cluster-10760.70530 | CGACGACGAGAAAGAAAGAGAG | AGCCCGAAGATGATGAAACC |
| Cluster-10760.67533 | GGGACGCAAGGTCGGAAT | GTCGAGTTCGGCGGGTTAG |
| Cluster-10760.67861 | GAAGGAAGGAACCCATCAAGT | CAAAATCCAGAGAAACGCAGA |
| Cluster-10760.6999 | CAGGAAGAAGGATGGGTAGTGT | TCGAGCTGTGGGAGTTGTATAA |
| Cluster-10760.70147 | TGTTGGTCTAGTTGGTGTTTCC | TTTTTGTTCGTAGTTCAGTGTGC |
| Cluster-10760.73033 | AAAAGATGAGAACGGCGAGAGT | AGGATGCGTAATAGGGGAAAAC |
| Cluster-10760.73468 | GATTCTTGGTTTTCTTCTGCTTT | CTACTTGATCTCGACGGTTTTG |
| Cluster-10760.73971 | ACTTTTGTGTTTTATTTGGGGG | TGAGGTGTTGTATTGGTGGTGA |
| Cluster-10760.79303 | ATCAAGCGGTAGTGGTGGC | GGTTTGGGGTAGGAAATGTGT |
| Cluster-10760.81770 | CTCTCTCTTACCATACCACCCC | GATTTCCAATACCCGCTACTC |
| Cluster-10760.89696 | TGTGTAAATAGGGGAGTGGGAG | AGGTGTCGGTTGGTAAAGTAGG |
| Cluster-10760.9534 | CCCAAATCCTATCAACTCCATC | GAATCCTTGCTTCCTTCACTCA |
| Cluster-1169.0 | CATTTTCCTCACATAGGCCAC | CACCCACCGATACATCATCAC |
| Cluster-13332.0 | TCGTCTAAGGGTTGTTGTTCAC | AGCAATTTCTCATTCTTGGGTAT |
| Cluster-17703.0 | GTTCTCGCACAAATAGAGCTACC | AAAGTCTTTCTCATTCCAACCATA |
| Cluster-22290.1 | ACACCAACGGCAACAATAGAAA | CAAGAGTAGAGGCATGGAACGA |
| Cluster-23414.0 | GGAGAGCGTTGTCAAGGAGAC | GGAACTAGTGAGGCGGGATG |
| Cluster-24277.0 | GGTGTTGGAGCAGAGGGC | GCATAGGGGGGAGGAGTGA |
| Cluster-2502.0 | GATAAGGAGATCTTAAGGGGGC | GAGAAGCAGTGTACGTGAGTCG |
| Cluster-26181.0 | TTTTGTAAGCCGAGTTGTGGTA | TCAGATCGTCATCGAAAGTGAT |
| Cluster-27488.0 | CCCCAAGGGTTTTTTATGTGT | ATGTCGGACTGTGGATTTTTTC |
| Cluster-6808.0 | ACGTCGGTAAAGCTCCAAAAG | GGTGCTAGGACCACCAAAAAT |
| Cluster-7497.0 | TAAGTTTATGCCGGGTGTATGTG | GTTGTTGTGAAAGGGTTGTGGA |
| 序号 No. | 基因名称 Gene name | Unigene | NCBI比对结果 Blast result | NCBI比对序列结果NCBI alignment sequence results | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 名称 Name | 长度 Length/bp | KEGG注释 KEGG annonation | 登录号 Accession number | cDNA长度 Length of cDNA | 氨基酸数量 Number of amino acids | 等电点 pI | 分子量 Mw | 脂肪系数 Aliphatic index | 疏水性 GRAVY | |||
| 1 | HtNAC7-1 | Cluster-10091.0 | 335 | NAC7 | HaNAC7 | XM_022167132.2 | 981 | 326 | 6.07 | 38 170.78 | 65.18 | -0.846 |
| 2 | HtNAC79-1 | Cluster-10760.100264 | 905 | NAC79 | HaNAC79 | XM_022157222.2 | 1 020 | 339 | 8.11 | 37 868.29 | 78.20 | -0.435 |
| Cluster-27142.0 | 428 | NAC79 | ||||||||||
| 3 | HtNAC53-1 | Cluster-10760.101731 | 777 | NAC53 | HaNAC53 | XM_022150409.2 | 1 020 | 577 | 4.48 | 65 141.09 | 72.31 | -0.600 |
| 4 | HtNAC2-1 | Cluster-10760.101956 | 320 | NAC2 | HaNAC2 | XM_022127653.2 | 876 | 291 | 6.46 | 33 637.00 | 57.97 | -0.713 |
| 5 | HtNAC92-1 | Cluster-10760.106687 | 572 | NAC92 | HaNAC92 | XM_022127585.2 | 1 032 | 343 | 7.15 | 39 502.76 | 64.81 | -0.592 |
| Cluster-10760.106688 | 344 | NAC92 | ||||||||||
| Cluster-10760.124019 | 791 | NAC92 | ||||||||||
| 6 | HtNAC2-2 | Cluster-10760.107212 | 393 | NAC2 | HaNAC2 | XM_022141099.2 | 849 | 282 | 6.40 | 32 308.70 | 64.33 | -0.598 |
| Cluster-10760.49834 | 231 | NAC2 | HaNAC2 | |||||||||
| Cluster-10760.64992 | 635 | NAC2 | HaNAC2 | |||||||||
| 7 | HtNAC2-3 | Cluster-10760.107377 | 246 | NAC2 | HaNAC2 | XM_022155734.2 | 843 | 280 | 6.12 | 32 152.13 | 56.43 | -0.727 |
| Cluster-10760.63798 | 767 | NAC2 | HaNAC2 | |||||||||
| Cluster-10760.76393 | 699 | NAC2 | HaNAC2 | |||||||||
| 8 | HtNAC79-2 | Cluster-10760.110871 | 761 | NAC79 | HaNAC79 | XM_022156995.2 | 879 | 292 | 6.56 | 33 705.19 | 56.71 | -0.749 |
| Cluster-10760.23420 | 602 | NAC79 | HaNAC79 | |||||||||
| Cluster-10760.28362 | 369 | NAC79 | HaNAC79 | |||||||||
| 9 | HtNAC90-1 | Cluster-10760.114800 | 953 | NAC90 | HaNAC90 | XM_022179099.2 | 705 | 234 | 8.69 | 27 398.75 | 57.52 | -0.810 |
| 10 | HtNAC67-1 | Cluster-10760.117836 | 776 | NAC67 | HaNAC67 | XM_022116617.2 | 909 | 302 | 5.52 | 34 585.60 | 59.40 | -0.874 |
| 11 | HtNAC67-2 | Cluster-10760.117837 | 626 | NAC67 | HaNAC67 | XM_022116619.2 | 885 | 294 | 5.30 | 33 866.10 | 70.61 | -0.712 |
| Cluster-10760.15238 | 385 | NAC67 | HaNAC67 | |||||||||
| 12 | HtNAC53-2 | Cluster-10760.121177 | 722 | NAC53 | HaNAC53 | XM_022137778.2 | 1 746 | 581 | 4.51 | 65 560.73 | 73.15 | -0.526 |
| Cluster-10760.46569 | 301 | NAC53 | HaNAC53 | |||||||||
| Cluster-10760.86661 | 1 179 | NAC53 | HaNAC53 | |||||||||
| 13 | HtNAC46 | Cluster-10760.125324 | 1 547 | NAC79 | HaNAC46 | XM_022170191.2 | 1 212 | 403 | 5.46 | 45 687.80 | 51.99 | -0.844 |
| Cluster-10760.125327 | 1 532 | NAC79 | HaNAC46 | |||||||||
| 14 | HtNAC87 | Cluster-10760.125325 | 890 | NAC100 | HaNAC87 | XM_022144895.2 | 1 158 | 385 | 5.63 | 43 335.23 | 56.16 | -0.683 |
| Cluster-10760.125326 | 1 216 | NAC100 | HaNAC87 | |||||||||
| 15 | HtNAC2-4 | Cluster-10760.12609 | 815 | NAC2 | HaNAC2 | XM_022127653.2 | 876 | 291 | 6.46 | 33 637.00 | 57.97 | -0.713 |
| Cluster-10760.73508 | 1 177 | NAC2 | HaNAC2 | |||||||||
| 16 | HtNAC82 | Cluster-10760.126290 | 541 | NAC82 | HaNAC82 | XM_022117092.2 | 1 137 | 378 | 4.40 | 41 245.86 | 71.69 | -0.343 |
| Cluster-10760.67517 | 1 573 | NAC82 | HaNAC82 | |||||||||
| 17 | HtNAC90-2 | Cluster-10760.129386 | 304 | NAC90 | HaNAC90 | XM_022149185.2 | 753 | 250 | 9.57 | 28 994.16 | 65.44 | -0.678 |
| 18 | HtNAC17-1 | Cluster-10760.130708 | 297 | NAC17 | HaNAC17 | XM_022184100.2 | 1 176 | 391 | 5.17 | 44 369.74 | 73.20 | -0.631 |
| 19 | HtNAC71-1 | Cluster-10760.130718 | 1 204 | NAC71 | HaNAC71 | XM_022173732.2 | 978 | 325 | 5.74 | 37 185.51 | 54.83 | -0.725 |
| 20 | HtNAC35-1 | Cluster-10760.132452 | 461 | NAC35 | HaNAC35 | XM_022172451.2 | 1 173 | 390 | 6.18 | 44 696.90 | 66.72 | -0.807 |
| Cluster-10760.26982 | 1 001 | NAC35 | HaNAC35 | |||||||||
| Cluster-10760.36796 | 1 484 | NAC35 | HaNAC35 | |||||||||
| 21 | HtNAC67-3 | Cluster-10760.133271 | 488 | NAC67 | HaNAC67 | XM_022116662.2 | 882 | 293 | 5.16 | 33 608.56 | 69.56 | -0.738 |
| 22 | HtNAC40 | Cluster-10760.134161 | 1 472 | NAC40 | HaNAC40 | XM_022146732.2 | 1 326 | 441 | 5.31 | 49 353.99 | 63.47 | -0.659 |
| 23 | HtNAC75-1 | Cluster-10760.135520 | 2 063 | NAC55 | HaNAC75 | XM_035980716.1 | 1 374 | 457 | 6.58 | 51 313.14 | 59.91 | -0.824 |
| 24 | HtNAC1 | Cluster-10760.135805 | 455 | NAC | HaNAC | XM_022164967.2 | 1 293 | 430 | 4.93 | 47 967.25 | 67.07 | -0.754 |
| 25 | HtNAC67-4 | Cluster-10760.137049 | 355 | NAC67 | HaNAC67 | XM_022170873.2 | 882 | 293 | 5.74 | 33 446.84 | 75.19 | -0.599 |
| Cluster-10760.137050 | 653 | NAC67 | HaNAC67 | |||||||||
| 26 | HtNAC86 | Cluster-10760.138345 | 577 | NAC86 | HaNAC86 | XM_022160342.2 | 1 869 | 622 | 5.16 | 70 271.98 | 64.50 | -0.686 |
| Cluster-10760.138346 | 1 161 | NAC86 | HaNAC86 | XM_035985121.1 | ||||||||
| 27 | HtNAC83-1 | Cluster-10760.16091 | 920 | NAC83 | HaNAC83 | XM_022120502.2 | 738 | 245 | 9.13 | 27 948.82 | 64.33 | -0.727 |
| Cluster-10760.41420 | 666 | NAC83 | HaNAC83 | |||||||||
| Cluster-10760.48444 | 518 | NAC83 | HaNAC83 | |||||||||
| Cluster-10760.93364 | 994 | NAC83 | HaNAC83 | |||||||||
| 28 | HtNAC21-1 | Cluster-10760.16732 | 459 | NAC21 | HaNAC21 | XM_022183930.2 | 963 | 320 | 5.90 | 36 454.97 | 66.06 | -0.610 |
| Cluster-10760.91122 | 577 | NAC | HaNAC21 | |||||||||
| 29 | HtNAC92-2 | Cluster-10760.17171 | 1 264 | NAC92 | HaNAC92 | XM_022167449.2 | 909 | 302 | 6.33 | 35 329.09 | 68.44 | -0.705 |
| 30 | HtNAC71-2 | Cluster-10760.19944 | 944 | NAC71 | HaNAC71 | XM_022146492.2 | 924 | 307 | 5.33 | 34 983.44 | 69.51 | -0.529 |
| Cluster-10760.29445 | 751 | NAC71 | HaNAC71 | |||||||||
| 31 32 | HtNAC22-1 HtNAC90-3 | Cluster-10760.26307 Cluster-10760.2853 | 1 377 680 | NAC35 NAC90 | HaNAC22 HaNAC90 | XM_022155988.2 XM_022152300.2 | 792 630 | 263 209 | 8.79 9.96 | 30 075.71 24 489.74 | 74.52 66.60 | -0.489 -0.842 |
| 32 | HtNAC90-3 | Cluster-10760.2853 | 680 | NAC90 | HaNAC90 | XM_022152300.2 | 630 | 209 | 9.96 | 24 489.74 | 66.60 | -0.842 |
| 33 | HtNAC50-1 | Cluster-10760.34552 | 1 348 | NAC16 | HaNAC50 | XM_022137779.2 | 1 338 | 445 | 7.57 | 50 237.19 | 72.09 | -0.576 |
| Cluster-12056.0 | 872 | NAC16 | HaNAC50 | |||||||||
| 34 | HtNAC26 | Cluster-10760.40188 | 475 | NAC26 | HaNAC26 | XM_022159224.2 | 780 | 259 | 4.90 | 29 021.66 | 78.26 | -0.369 |
| Cluster-10760.40189 | 923 | NAC26 | HaNAC | |||||||||
| 35 | HtNAC83-2 | Cluster-10760.41713 | 1 177 | NAC83 | HaNAC83 | XM_022183428.2 | 816 | 271 | 9.25 | 31 044.44 | 68.63 | -0.608 |
| 36 | HtNAC73 | Cluster-10760.4894 | 1 101 | NAC73 | HaNAC73 | XM_022179643.2 | 870 | 289 | 8.77 | 32 571.68 | 64.71 | -0.785 |
| Cluster-10760.54303 | 392 | NAC73 | HaNAC73 | |||||||||
| 37 | HtNAC50-2 | Cluster-10760.49107 | 995 | NAC79 | HaNAC50 | XM_022166965.2 | 1 389 | 462 | 5.35 | 51 849.35 | 64.96 | -0.883 |
| Cluster-10760.53282 | 820 | NAC79 | HaNAC50 | |||||||||
| Cluster-10760.86831 | 1 573 | NAC79 | HaNAC50 | |||||||||
| 38 | HtNAC83-3 | Cluster-10760.53392 | 937 | NAC83 | HaNAC83 | XM_022167085.2 | 627 | 208 | 9.36 | 23 438.62 | 67.88 | -0.646 |
| 39 | HtNAC50-3 | Cluster-10760.53462 | 221 | NAC78 | HaNAC50 | XM_022121208.2 | 1 134 | 377 | 5.27 | 42 391.52 | 65.76 | -0.685 |
| Cluster-10760.66624 | 600 | NAC78 | HaNAC50 | |||||||||
| Cluster-10760.74589 | 1 339 | NAC78 | HaNAC50 | |||||||||
| 40 | HtNAC75-2 | Cluster-10760.54233 | 1 259 | NAC86 | HaNAC75 | XM_022125233.2 | 1 326 | 441 | 6.33 | 50 307.19 | 60.50 | -0.866 |
| 41 | HtNAC90-4 | Cluster-10760.54351 | 1 016 | NAC90 | HaNAC90 | XM_022153711.2 | 570 | 189 | 8.44 | 21 699.61 | 66.46 | -0.590 |
| Cluster-6868.0 | 284 | NAC90 | HaNAC90 | |||||||||
| 42 | HtNAC21-2 | Cluster-10760.54670 | 1 525 | NAC21 | HaNAC21 | XM_022122480.2 | 933 | 310 | 8.02 | 35 217.62 | 68.23 | -0.570 |
| 43 | HtNAC90-5 | Cluster-10760.57561 | 609 | NAC90 | HaNAC90 | XM_022149185.2 | 753 | 250 | 9.57 | 28 994.16 | 65.44 | -0.678 |
| 44 | HtNAC50-4 | Cluster-10760.60147 | 986 | NAC101 | HaNAC50 | XM_022178589.2 | 993 | 330 | 5.23 | 36 710.04 | 65.61 | -0.584 |
| 45 | HtNAC17-2 | Cluster-10760.63247 | 1 840 | NAC17 | HaNAC17 | XM_022184099.2 | 1 494 | 497 | 5.13 | 55 812.64 | 75.05 | -0.445 |
| 46 | HtNAC53-3 | Cluster-10760.63290 | 1 194 | NAC53 | HaNAC53 | XM_022150409.2 | 1 734 | 577 | 4.48 | 65 141.09 | 72.31 | -0.600 |
| Cluster-10760.76166 | 591 | NAC53 | HaNAC53 | XM_022150409.2 | 1 734 | 577 | 4.48 | 65 141.09 | 72.31 | -0.600 | ||
| 47 | HtNAC83-4 | Cluster-10760.65303 | 711 | NAC83 | HaNAC83 | XM_022120532.2 | 408 | 135 | 9.24 | 15 288.34 | 61.33 | -0.847 |
| Cluster-10760.69351 | 825 | NAC83 | HaNAC83 | XM_022120532.2 | ||||||||
| 48 | HtNAC83-5 | Cluster-10760.66194 | 238 | NAC83 | HaNAC83 | XM_022141545.2 | 747 | 248 | 9.17 | 28 139.04 | 66.73 | -0.717 |
| Cluster-10760.69247 | 451 | NAC83 | HaNAC83 | XM_022141545.2 | ||||||||
| Cluster-10760.70530 | 1 133 | NAC83 | HaNAC83 | XM_022141545.2 | ||||||||
| Cluster-10760.80917 | 218 | NAC83 | HaNAC83 | XM_022141545.2 | ||||||||
| 49 | HtNACJA2L | Cluster-10760.67533 | 1503 | NAC72 | HaNACJA2L | XM_022173110.2 | 930 | 309 | 7.68 | 34 968.28 | 60.23 | -0.714 |
| 50 | HtNAC78 | Cluster-10760.67861 | 1877 | NAC78 | HaNAC78 | XM_022121205.2 | 1 611 | 536 | 4.67 | 59 876.02 | 73.47 | -0.416 |
| 51 | HtNAC37 | Cluster-10760.6999 | 1302 | NAC37 | HaNAC37 | XM_022153995.2 | 1 062 | 353 | 5.34 | 40 758.58 | 64.33 | -0.738 |
| 52 | HtNAC2-5 | Cluster-10760.70147 | 1 163 | NAC83 | HaNAC2 | XM_035983703.1 | 1 359 | 452 | 4.91 | 49 639.65 | 68.85 | -0.549 |
| Cluster-2535.0 | 233 | NAC83 | HaNAC2 | XM_035983703.1 | ||||||||
| 53 | HtNAC62 | Cluster-10760.73033 | 1 257 | NAC91 | HaNAC62 | XM_022131917.2 | 1 653 | 550 | 5.17 | 61 509.66 | 69.04 | -0.643 |
| 54 | HtNAC17-3 | Cluster-10760.73468 | 1 406 | NAC17 | HaNAC17 | XM_022131488.2 | 1 545 | 514 | 4.79 | 57 445.07 | 72.65 | -0.531 |
| Cluster-10760.76418 | 527 | NAC17 | HaNAC17 | XM_022131488.2 | ||||||||
| 55 | HtNAC72 | Cluster-10760.73971 | 961 | NAC72 | HaNAC72 | XM_022183015.2 | 663 | 220 | 8.89 | 24 874.64 | 76.59 | -0.349 |
| 56 | HtNAC100-1 | Cluster-10760.79303 | 1 571 | NAC100 | HaNAC100 | XM_022183364.2 | 1 020 | 339 | 7.68 | 37 932.65 | 61.80 | -0.664 |
| 57 | HtNAC104 | Cluster-10760.81770 | 867 | NAC104 | HaNAC104 | XM_022132807.2 | 597 | 198 | 4.93 | 22 724.18 | 63.94 | -0.684 |
| 58 | HtNAC83-6 | Cluster-10760.89696 | 1 538 | NAC83 | HaNAC83 | XM_022155604.2 | 801 | 266 | 8.82 | 30 509.31 | 63.72 | -0.763 |
| 59 | HtNAC35-2 | Cluster-10760.9533 | 277 | NAC35 | HaNAC35 | XM_022131569.2 | 1 107 | 368 | 7.70 | 42 393.56 | 67.58 | -0.826 |
| Cluster-10760.9534 | 2 102 | NAC35 | HaNAC35 | XM_022131569.2 | ||||||||
| 60 | HtNAC94 | Cluster-1169.0 | 1 628 | NAC94 | HaNAC94 | XM_022127636.2 | 1 056 | 351 | 6.76 | 40 010.08 | 60.85 | -0.630 |
| 61 | HtNAC7-2 | Cluster-13332.0 | 862 | NAC7 | HaNAC7 | XM_022144664.2 | 987 | 328 | 6.28 | 38 409.01 | 62.13 | -0.872 |
| Cluster-26650.1 | 352 | NAC | HaNAC7 | XM_022144664.2 | ||||||||
| 62 | HtNAC7-3 | Cluster-17703.0 | 749 | NAC7 | HaNAC7 | XM_022118485.2 | 975 | 324 | 5.40 | 37 795.32 | 67.96 | -0.734 |
| 63 | HtNAC50-5 | Cluster-22290.0 | 441 | NAC78 | HaNAC50 | XM_022150408.2 | 1 194 | 397 | 5.57 | 44 738.48 | 72.90 | -0.562 |
| Cluster-22290.1 | 1 402 | NAC78 | HaNAC50 | XM_022150408.2 | ||||||||
| 64 | HtNAC90-6 | Cluster-23414.0 | 424 | NAC90 | HaNAC90 | XM_035981818.1 | 759 | 252 | 8.59 | 29 166.73 | 62.26 | -0.754 |
| 65 | HtNAC22-2 | Cluster-24277.0 | 780 | NAC35 | HaNAC22 | XM_022165755.2 | 822 | 273 | 9.00 | 31 328.00 | 73.59 | -0.465 |
| 66 | Ht-NAC100-2 | Cluster-2502.0 | 1 172 | NAC100 | HaNAC100 | XM_022161816.2 | 933 | 310 | 6.80 | 35 062.82 | 59.42 | -0.606 |
| 67 | HtNAC83-7 | Cluster-26181.0 | 644 | NAC83 | HaNAC83 | XM_022126764.2 | 684 | 227 | 8.46 | 26 016.74 | 76.43 | -0.546 |
| 68 | HtNAC76 | Cluster-27488.0 | 701 | NAC76 | HaNAC76 | XM_022162486.2 | 927 | 308 | 6.03 | 35 898.38 | 66.75 | -0.735 |
| 69 | HtNACJA2 | Cluster-6808.0 | 318 | NAC19 | HaNACJA2 | XM_022135152.1 | 684 | 227 | 8.75 | 26 250.02 | 61.76 | -0.554 |
| 70 | HtNAC90-7 | Cluster-7497.0 | 1 040 | NAC90 | HaNAC90 | XM_022179077.2 | 774 | 257 | 6.21 | 30 100.55 | 54.20 | -0.906 |
表2 HtNAC基因理化性质分析
Table 2 Physicochemical properties of HtNAC gene
| 序号 No. | 基因名称 Gene name | Unigene | NCBI比对结果 Blast result | NCBI比对序列结果NCBI alignment sequence results | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 名称 Name | 长度 Length/bp | KEGG注释 KEGG annonation | 登录号 Accession number | cDNA长度 Length of cDNA | 氨基酸数量 Number of amino acids | 等电点 pI | 分子量 Mw | 脂肪系数 Aliphatic index | 疏水性 GRAVY | |||
| 1 | HtNAC7-1 | Cluster-10091.0 | 335 | NAC7 | HaNAC7 | XM_022167132.2 | 981 | 326 | 6.07 | 38 170.78 | 65.18 | -0.846 |
| 2 | HtNAC79-1 | Cluster-10760.100264 | 905 | NAC79 | HaNAC79 | XM_022157222.2 | 1 020 | 339 | 8.11 | 37 868.29 | 78.20 | -0.435 |
| Cluster-27142.0 | 428 | NAC79 | ||||||||||
| 3 | HtNAC53-1 | Cluster-10760.101731 | 777 | NAC53 | HaNAC53 | XM_022150409.2 | 1 020 | 577 | 4.48 | 65 141.09 | 72.31 | -0.600 |
| 4 | HtNAC2-1 | Cluster-10760.101956 | 320 | NAC2 | HaNAC2 | XM_022127653.2 | 876 | 291 | 6.46 | 33 637.00 | 57.97 | -0.713 |
| 5 | HtNAC92-1 | Cluster-10760.106687 | 572 | NAC92 | HaNAC92 | XM_022127585.2 | 1 032 | 343 | 7.15 | 39 502.76 | 64.81 | -0.592 |
| Cluster-10760.106688 | 344 | NAC92 | ||||||||||
| Cluster-10760.124019 | 791 | NAC92 | ||||||||||
| 6 | HtNAC2-2 | Cluster-10760.107212 | 393 | NAC2 | HaNAC2 | XM_022141099.2 | 849 | 282 | 6.40 | 32 308.70 | 64.33 | -0.598 |
| Cluster-10760.49834 | 231 | NAC2 | HaNAC2 | |||||||||
| Cluster-10760.64992 | 635 | NAC2 | HaNAC2 | |||||||||
| 7 | HtNAC2-3 | Cluster-10760.107377 | 246 | NAC2 | HaNAC2 | XM_022155734.2 | 843 | 280 | 6.12 | 32 152.13 | 56.43 | -0.727 |
| Cluster-10760.63798 | 767 | NAC2 | HaNAC2 | |||||||||
| Cluster-10760.76393 | 699 | NAC2 | HaNAC2 | |||||||||
| 8 | HtNAC79-2 | Cluster-10760.110871 | 761 | NAC79 | HaNAC79 | XM_022156995.2 | 879 | 292 | 6.56 | 33 705.19 | 56.71 | -0.749 |
| Cluster-10760.23420 | 602 | NAC79 | HaNAC79 | |||||||||
| Cluster-10760.28362 | 369 | NAC79 | HaNAC79 | |||||||||
| 9 | HtNAC90-1 | Cluster-10760.114800 | 953 | NAC90 | HaNAC90 | XM_022179099.2 | 705 | 234 | 8.69 | 27 398.75 | 57.52 | -0.810 |
| 10 | HtNAC67-1 | Cluster-10760.117836 | 776 | NAC67 | HaNAC67 | XM_022116617.2 | 909 | 302 | 5.52 | 34 585.60 | 59.40 | -0.874 |
| 11 | HtNAC67-2 | Cluster-10760.117837 | 626 | NAC67 | HaNAC67 | XM_022116619.2 | 885 | 294 | 5.30 | 33 866.10 | 70.61 | -0.712 |
| Cluster-10760.15238 | 385 | NAC67 | HaNAC67 | |||||||||
| 12 | HtNAC53-2 | Cluster-10760.121177 | 722 | NAC53 | HaNAC53 | XM_022137778.2 | 1 746 | 581 | 4.51 | 65 560.73 | 73.15 | -0.526 |
| Cluster-10760.46569 | 301 | NAC53 | HaNAC53 | |||||||||
| Cluster-10760.86661 | 1 179 | NAC53 | HaNAC53 | |||||||||
| 13 | HtNAC46 | Cluster-10760.125324 | 1 547 | NAC79 | HaNAC46 | XM_022170191.2 | 1 212 | 403 | 5.46 | 45 687.80 | 51.99 | -0.844 |
| Cluster-10760.125327 | 1 532 | NAC79 | HaNAC46 | |||||||||
| 14 | HtNAC87 | Cluster-10760.125325 | 890 | NAC100 | HaNAC87 | XM_022144895.2 | 1 158 | 385 | 5.63 | 43 335.23 | 56.16 | -0.683 |
| Cluster-10760.125326 | 1 216 | NAC100 | HaNAC87 | |||||||||
| 15 | HtNAC2-4 | Cluster-10760.12609 | 815 | NAC2 | HaNAC2 | XM_022127653.2 | 876 | 291 | 6.46 | 33 637.00 | 57.97 | -0.713 |
| Cluster-10760.73508 | 1 177 | NAC2 | HaNAC2 | |||||||||
| 16 | HtNAC82 | Cluster-10760.126290 | 541 | NAC82 | HaNAC82 | XM_022117092.2 | 1 137 | 378 | 4.40 | 41 245.86 | 71.69 | -0.343 |
| Cluster-10760.67517 | 1 573 | NAC82 | HaNAC82 | |||||||||
| 17 | HtNAC90-2 | Cluster-10760.129386 | 304 | NAC90 | HaNAC90 | XM_022149185.2 | 753 | 250 | 9.57 | 28 994.16 | 65.44 | -0.678 |
| 18 | HtNAC17-1 | Cluster-10760.130708 | 297 | NAC17 | HaNAC17 | XM_022184100.2 | 1 176 | 391 | 5.17 | 44 369.74 | 73.20 | -0.631 |
| 19 | HtNAC71-1 | Cluster-10760.130718 | 1 204 | NAC71 | HaNAC71 | XM_022173732.2 | 978 | 325 | 5.74 | 37 185.51 | 54.83 | -0.725 |
| 20 | HtNAC35-1 | Cluster-10760.132452 | 461 | NAC35 | HaNAC35 | XM_022172451.2 | 1 173 | 390 | 6.18 | 44 696.90 | 66.72 | -0.807 |
| Cluster-10760.26982 | 1 001 | NAC35 | HaNAC35 | |||||||||
| Cluster-10760.36796 | 1 484 | NAC35 | HaNAC35 | |||||||||
| 21 | HtNAC67-3 | Cluster-10760.133271 | 488 | NAC67 | HaNAC67 | XM_022116662.2 | 882 | 293 | 5.16 | 33 608.56 | 69.56 | -0.738 |
| 22 | HtNAC40 | Cluster-10760.134161 | 1 472 | NAC40 | HaNAC40 | XM_022146732.2 | 1 326 | 441 | 5.31 | 49 353.99 | 63.47 | -0.659 |
| 23 | HtNAC75-1 | Cluster-10760.135520 | 2 063 | NAC55 | HaNAC75 | XM_035980716.1 | 1 374 | 457 | 6.58 | 51 313.14 | 59.91 | -0.824 |
| 24 | HtNAC1 | Cluster-10760.135805 | 455 | NAC | HaNAC | XM_022164967.2 | 1 293 | 430 | 4.93 | 47 967.25 | 67.07 | -0.754 |
| 25 | HtNAC67-4 | Cluster-10760.137049 | 355 | NAC67 | HaNAC67 | XM_022170873.2 | 882 | 293 | 5.74 | 33 446.84 | 75.19 | -0.599 |
| Cluster-10760.137050 | 653 | NAC67 | HaNAC67 | |||||||||
| 26 | HtNAC86 | Cluster-10760.138345 | 577 | NAC86 | HaNAC86 | XM_022160342.2 | 1 869 | 622 | 5.16 | 70 271.98 | 64.50 | -0.686 |
| Cluster-10760.138346 | 1 161 | NAC86 | HaNAC86 | XM_035985121.1 | ||||||||
| 27 | HtNAC83-1 | Cluster-10760.16091 | 920 | NAC83 | HaNAC83 | XM_022120502.2 | 738 | 245 | 9.13 | 27 948.82 | 64.33 | -0.727 |
| Cluster-10760.41420 | 666 | NAC83 | HaNAC83 | |||||||||
| Cluster-10760.48444 | 518 | NAC83 | HaNAC83 | |||||||||
| Cluster-10760.93364 | 994 | NAC83 | HaNAC83 | |||||||||
| 28 | HtNAC21-1 | Cluster-10760.16732 | 459 | NAC21 | HaNAC21 | XM_022183930.2 | 963 | 320 | 5.90 | 36 454.97 | 66.06 | -0.610 |
| Cluster-10760.91122 | 577 | NAC | HaNAC21 | |||||||||
| 29 | HtNAC92-2 | Cluster-10760.17171 | 1 264 | NAC92 | HaNAC92 | XM_022167449.2 | 909 | 302 | 6.33 | 35 329.09 | 68.44 | -0.705 |
| 30 | HtNAC71-2 | Cluster-10760.19944 | 944 | NAC71 | HaNAC71 | XM_022146492.2 | 924 | 307 | 5.33 | 34 983.44 | 69.51 | -0.529 |
| Cluster-10760.29445 | 751 | NAC71 | HaNAC71 | |||||||||
| 31 32 | HtNAC22-1 HtNAC90-3 | Cluster-10760.26307 Cluster-10760.2853 | 1 377 680 | NAC35 NAC90 | HaNAC22 HaNAC90 | XM_022155988.2 XM_022152300.2 | 792 630 | 263 209 | 8.79 9.96 | 30 075.71 24 489.74 | 74.52 66.60 | -0.489 -0.842 |
| 32 | HtNAC90-3 | Cluster-10760.2853 | 680 | NAC90 | HaNAC90 | XM_022152300.2 | 630 | 209 | 9.96 | 24 489.74 | 66.60 | -0.842 |
| 33 | HtNAC50-1 | Cluster-10760.34552 | 1 348 | NAC16 | HaNAC50 | XM_022137779.2 | 1 338 | 445 | 7.57 | 50 237.19 | 72.09 | -0.576 |
| Cluster-12056.0 | 872 | NAC16 | HaNAC50 | |||||||||
| 34 | HtNAC26 | Cluster-10760.40188 | 475 | NAC26 | HaNAC26 | XM_022159224.2 | 780 | 259 | 4.90 | 29 021.66 | 78.26 | -0.369 |
| Cluster-10760.40189 | 923 | NAC26 | HaNAC | |||||||||
| 35 | HtNAC83-2 | Cluster-10760.41713 | 1 177 | NAC83 | HaNAC83 | XM_022183428.2 | 816 | 271 | 9.25 | 31 044.44 | 68.63 | -0.608 |
| 36 | HtNAC73 | Cluster-10760.4894 | 1 101 | NAC73 | HaNAC73 | XM_022179643.2 | 870 | 289 | 8.77 | 32 571.68 | 64.71 | -0.785 |
| Cluster-10760.54303 | 392 | NAC73 | HaNAC73 | |||||||||
| 37 | HtNAC50-2 | Cluster-10760.49107 | 995 | NAC79 | HaNAC50 | XM_022166965.2 | 1 389 | 462 | 5.35 | 51 849.35 | 64.96 | -0.883 |
| Cluster-10760.53282 | 820 | NAC79 | HaNAC50 | |||||||||
| Cluster-10760.86831 | 1 573 | NAC79 | HaNAC50 | |||||||||
| 38 | HtNAC83-3 | Cluster-10760.53392 | 937 | NAC83 | HaNAC83 | XM_022167085.2 | 627 | 208 | 9.36 | 23 438.62 | 67.88 | -0.646 |
| 39 | HtNAC50-3 | Cluster-10760.53462 | 221 | NAC78 | HaNAC50 | XM_022121208.2 | 1 134 | 377 | 5.27 | 42 391.52 | 65.76 | -0.685 |
| Cluster-10760.66624 | 600 | NAC78 | HaNAC50 | |||||||||
| Cluster-10760.74589 | 1 339 | NAC78 | HaNAC50 | |||||||||
| 40 | HtNAC75-2 | Cluster-10760.54233 | 1 259 | NAC86 | HaNAC75 | XM_022125233.2 | 1 326 | 441 | 6.33 | 50 307.19 | 60.50 | -0.866 |
| 41 | HtNAC90-4 | Cluster-10760.54351 | 1 016 | NAC90 | HaNAC90 | XM_022153711.2 | 570 | 189 | 8.44 | 21 699.61 | 66.46 | -0.590 |
| Cluster-6868.0 | 284 | NAC90 | HaNAC90 | |||||||||
| 42 | HtNAC21-2 | Cluster-10760.54670 | 1 525 | NAC21 | HaNAC21 | XM_022122480.2 | 933 | 310 | 8.02 | 35 217.62 | 68.23 | -0.570 |
| 43 | HtNAC90-5 | Cluster-10760.57561 | 609 | NAC90 | HaNAC90 | XM_022149185.2 | 753 | 250 | 9.57 | 28 994.16 | 65.44 | -0.678 |
| 44 | HtNAC50-4 | Cluster-10760.60147 | 986 | NAC101 | HaNAC50 | XM_022178589.2 | 993 | 330 | 5.23 | 36 710.04 | 65.61 | -0.584 |
| 45 | HtNAC17-2 | Cluster-10760.63247 | 1 840 | NAC17 | HaNAC17 | XM_022184099.2 | 1 494 | 497 | 5.13 | 55 812.64 | 75.05 | -0.445 |
| 46 | HtNAC53-3 | Cluster-10760.63290 | 1 194 | NAC53 | HaNAC53 | XM_022150409.2 | 1 734 | 577 | 4.48 | 65 141.09 | 72.31 | -0.600 |
| Cluster-10760.76166 | 591 | NAC53 | HaNAC53 | XM_022150409.2 | 1 734 | 577 | 4.48 | 65 141.09 | 72.31 | -0.600 | ||
| 47 | HtNAC83-4 | Cluster-10760.65303 | 711 | NAC83 | HaNAC83 | XM_022120532.2 | 408 | 135 | 9.24 | 15 288.34 | 61.33 | -0.847 |
| Cluster-10760.69351 | 825 | NAC83 | HaNAC83 | XM_022120532.2 | ||||||||
| 48 | HtNAC83-5 | Cluster-10760.66194 | 238 | NAC83 | HaNAC83 | XM_022141545.2 | 747 | 248 | 9.17 | 28 139.04 | 66.73 | -0.717 |
| Cluster-10760.69247 | 451 | NAC83 | HaNAC83 | XM_022141545.2 | ||||||||
| Cluster-10760.70530 | 1 133 | NAC83 | HaNAC83 | XM_022141545.2 | ||||||||
| Cluster-10760.80917 | 218 | NAC83 | HaNAC83 | XM_022141545.2 | ||||||||
| 49 | HtNACJA2L | Cluster-10760.67533 | 1503 | NAC72 | HaNACJA2L | XM_022173110.2 | 930 | 309 | 7.68 | 34 968.28 | 60.23 | -0.714 |
| 50 | HtNAC78 | Cluster-10760.67861 | 1877 | NAC78 | HaNAC78 | XM_022121205.2 | 1 611 | 536 | 4.67 | 59 876.02 | 73.47 | -0.416 |
| 51 | HtNAC37 | Cluster-10760.6999 | 1302 | NAC37 | HaNAC37 | XM_022153995.2 | 1 062 | 353 | 5.34 | 40 758.58 | 64.33 | -0.738 |
| 52 | HtNAC2-5 | Cluster-10760.70147 | 1 163 | NAC83 | HaNAC2 | XM_035983703.1 | 1 359 | 452 | 4.91 | 49 639.65 | 68.85 | -0.549 |
| Cluster-2535.0 | 233 | NAC83 | HaNAC2 | XM_035983703.1 | ||||||||
| 53 | HtNAC62 | Cluster-10760.73033 | 1 257 | NAC91 | HaNAC62 | XM_022131917.2 | 1 653 | 550 | 5.17 | 61 509.66 | 69.04 | -0.643 |
| 54 | HtNAC17-3 | Cluster-10760.73468 | 1 406 | NAC17 | HaNAC17 | XM_022131488.2 | 1 545 | 514 | 4.79 | 57 445.07 | 72.65 | -0.531 |
| Cluster-10760.76418 | 527 | NAC17 | HaNAC17 | XM_022131488.2 | ||||||||
| 55 | HtNAC72 | Cluster-10760.73971 | 961 | NAC72 | HaNAC72 | XM_022183015.2 | 663 | 220 | 8.89 | 24 874.64 | 76.59 | -0.349 |
| 56 | HtNAC100-1 | Cluster-10760.79303 | 1 571 | NAC100 | HaNAC100 | XM_022183364.2 | 1 020 | 339 | 7.68 | 37 932.65 | 61.80 | -0.664 |
| 57 | HtNAC104 | Cluster-10760.81770 | 867 | NAC104 | HaNAC104 | XM_022132807.2 | 597 | 198 | 4.93 | 22 724.18 | 63.94 | -0.684 |
| 58 | HtNAC83-6 | Cluster-10760.89696 | 1 538 | NAC83 | HaNAC83 | XM_022155604.2 | 801 | 266 | 8.82 | 30 509.31 | 63.72 | -0.763 |
| 59 | HtNAC35-2 | Cluster-10760.9533 | 277 | NAC35 | HaNAC35 | XM_022131569.2 | 1 107 | 368 | 7.70 | 42 393.56 | 67.58 | -0.826 |
| Cluster-10760.9534 | 2 102 | NAC35 | HaNAC35 | XM_022131569.2 | ||||||||
| 60 | HtNAC94 | Cluster-1169.0 | 1 628 | NAC94 | HaNAC94 | XM_022127636.2 | 1 056 | 351 | 6.76 | 40 010.08 | 60.85 | -0.630 |
| 61 | HtNAC7-2 | Cluster-13332.0 | 862 | NAC7 | HaNAC7 | XM_022144664.2 | 987 | 328 | 6.28 | 38 409.01 | 62.13 | -0.872 |
| Cluster-26650.1 | 352 | NAC | HaNAC7 | XM_022144664.2 | ||||||||
| 62 | HtNAC7-3 | Cluster-17703.0 | 749 | NAC7 | HaNAC7 | XM_022118485.2 | 975 | 324 | 5.40 | 37 795.32 | 67.96 | -0.734 |
| 63 | HtNAC50-5 | Cluster-22290.0 | 441 | NAC78 | HaNAC50 | XM_022150408.2 | 1 194 | 397 | 5.57 | 44 738.48 | 72.90 | -0.562 |
| Cluster-22290.1 | 1 402 | NAC78 | HaNAC50 | XM_022150408.2 | ||||||||
| 64 | HtNAC90-6 | Cluster-23414.0 | 424 | NAC90 | HaNAC90 | XM_035981818.1 | 759 | 252 | 8.59 | 29 166.73 | 62.26 | -0.754 |
| 65 | HtNAC22-2 | Cluster-24277.0 | 780 | NAC35 | HaNAC22 | XM_022165755.2 | 822 | 273 | 9.00 | 31 328.00 | 73.59 | -0.465 |
| 66 | Ht-NAC100-2 | Cluster-2502.0 | 1 172 | NAC100 | HaNAC100 | XM_022161816.2 | 933 | 310 | 6.80 | 35 062.82 | 59.42 | -0.606 |
| 67 | HtNAC83-7 | Cluster-26181.0 | 644 | NAC83 | HaNAC83 | XM_022126764.2 | 684 | 227 | 8.46 | 26 016.74 | 76.43 | -0.546 |
| 68 | HtNAC76 | Cluster-27488.0 | 701 | NAC76 | HaNAC76 | XM_022162486.2 | 927 | 308 | 6.03 | 35 898.38 | 66.75 | -0.735 |
| 69 | HtNACJA2 | Cluster-6808.0 | 318 | NAC19 | HaNACJA2 | XM_022135152.1 | 684 | 227 | 8.75 | 26 250.02 | 61.76 | -0.554 |
| 70 | HtNAC90-7 | Cluster-7497.0 | 1 040 | NAC90 | HaNAC90 | XM_022179077.2 | 774 | 257 | 6.21 | 30 100.55 | 54.20 | -0.906 |
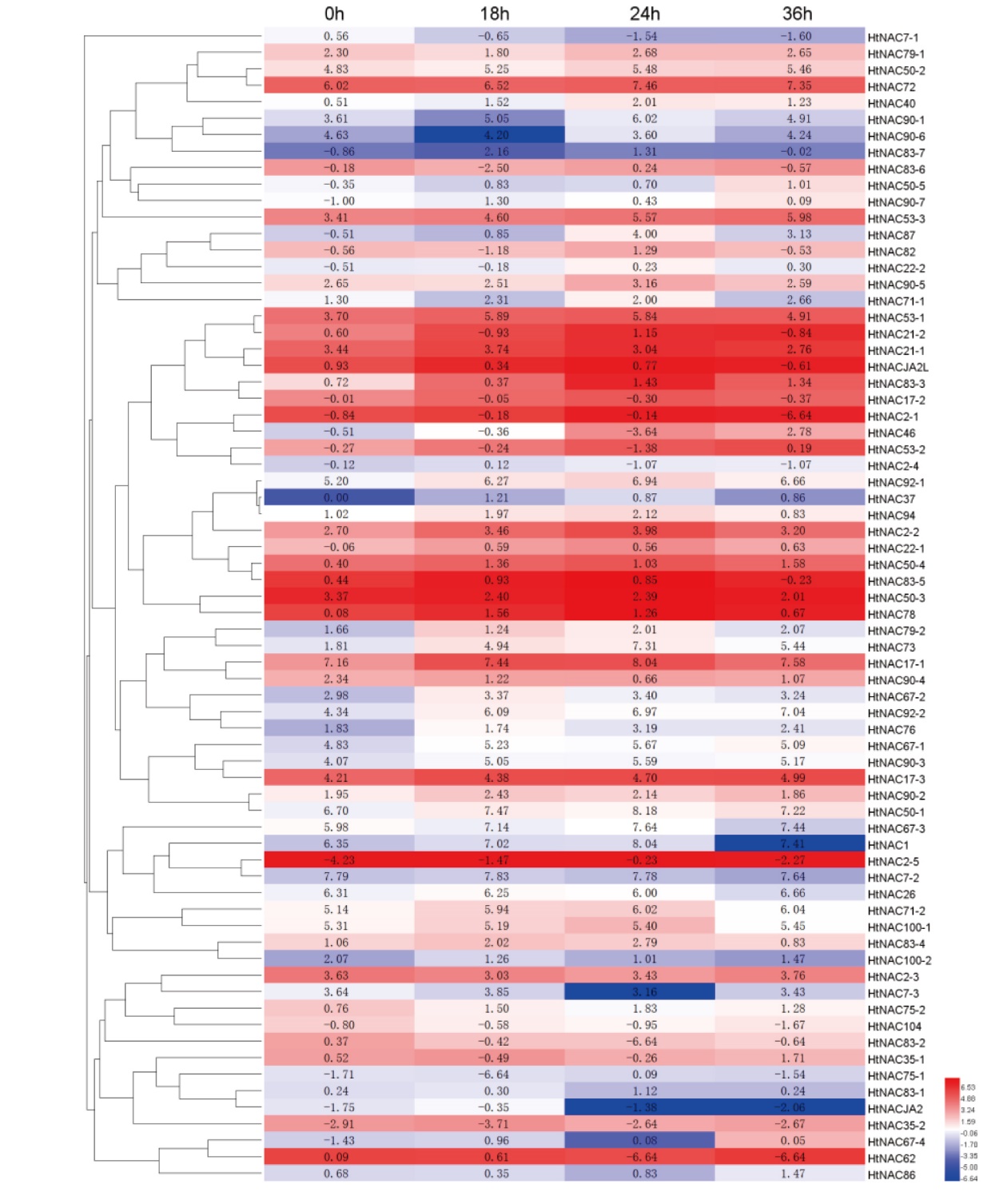
图5 70个HtNAC基因在PEG-6000模拟干旱胁迫的菊芋叶片中的表达水平热图。数字表示log2PFKM值
Fig. 5 Expression heatmap of 70 HtNAC genes under polyethylene glycol-simulated drought stress in H. tuberosus L. leaves. Number indicates log2PFKM value

图6 21个HtNAC基因在干旱胁迫下菊芋根中的表达情况 不同字母表示0.05显著水平下不同处理时期的差异性
Fig. 6 Expressions of 21 HtNAC genes in H. tuberosus L. roots under drought stress Different letters indicate the difference between different treatment periods at the significant level of 0.05
| [1] |
Long XH, Shao HB, Liu L, et al. Jerusalem artichoke: a sustainable biomass feedstock for biorefinery[J]. Renew Sustain Energy Rev, 2016, 54: 1382-1388.
doi: 10.1016/j.rser.2015.10.063 URL |
| [2] | Kays SJ, Nottingham SF. Biology and chemistry of jerusalem artichoke: Helianthus tuberosus L.[M]. Boca Raton: CRC Press, 2007. |
| [3] |
Lachman J, Kays SJ, Nottingham SF. Biology and chemistry of Jerusalem artichoke Helianthus tuberosus L[J]. Biol Plant, 2008, 52(3): 492.
doi: 10.1007/s10535-008-0095-3 URL |
| [4] |
Zhao ML, Ren YJ, Wei W, et al. Metabolite analysis of Jerusalem artichoke(Helianthus tuberosus L.) seedlings in response to polyethylene glycol-simulated drought stress[J]. Int J Mol Sci, 2021, 22(7): 3294.
doi: 10.3390/ijms22073294 URL |
| [5] |
Long XH, Chi JH, Liu L, et al. Effect of seawater stress on physiological and biochemical responses of five Jerusalem artichoke ecotypes[J]. Pedosphere, 2009, 19(2): 208-216.
doi: 10.1016/S1002-0160(09)60110-7 URL |
| [6] |
Wright L, Wrench P, Hinde RW, et al. Proline accumulation in tubers of Jerusalem artichoke[J]. Funct Plant Biol, 1977, 4(1): 51.
doi: 10.1071/PP9770051 URL |
| [7] |
Barta J, Pátkai G. Chemical composition and storability of Jerusalem artichoke tubers[J]. Acta Aliment, 2007, 36(2): 257-267.
doi: 10.1556/AAlim.36.2007.2.13 URL |
| [8] |
Long XH, Huang ZR, Zhang ZH, et al. Seawater stress differentially affects germination, growth, photosynthesis, and ion concentration in genotypes of Jerusalem artichoke(Helianthus tuberosus L.)[J]. J Plant Growth Regul, 2010, 29(2): 223-231.
doi: 10.1007/s00344-009-9125-4 URL |
| [9] |
Long XH, Huang ZR, Huang YL, et al. Response of two Jerusalem artichoke(Helianthus tuberosus)cultivars differing in tolerance to salt treatment[J]. Pedosphere, 2010, 20(4): 515-524.
doi: 10.1016/S1002-0160(10)60041-0 URL |
| [10] |
Jung WY, Lee SS, Kim CW, et al. RNA-seq analysis and de novo transcriptome assembly of Jerusalem artichoke(Helianthus tuberosus Linne)[J]. PLoS One, 2014, 9(11): e111982.
doi: 10.1371/journal.pone.0111982 URL |
| [11] |
Aida M, Ishida T, Fukaki H, et al. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant[J]. Plant Cell, 1997, 9(6): 841-857.
doi: 10.1105/tpc.9.6.841 pmid: 9212461 |
| [12] |
Puranik S, Sahu PP, Srivastava PS, et al. NAC proteins: regulation and role in stress tolerance[J]. Trends Plant Sci, 2012, 17(6): 369-381.
doi: 10.1016/j.tplants.2012.02.004 pmid: 22445067 |
| [13] |
Du MM, Zhai QZ, Deng L, et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack[J]. Plant Cell, 2014, 26(7): 3167-3184.
doi: 10.1105/tpc.114.128272 URL |
| [14] |
Giovannoni JJ. Fruit ripening mutants yield insights into ripening control[J]. Curr Opin Plant Biol, 2007, 10(3): 283-289.
doi: 10.1016/j.pbi.2007.04.008 pmid: 17442612 |
| [15] |
Hao YJ, Wei W, Song QX, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants[J]. Plant J, 2011, 68(2): 302-313.
doi: 10.1111/j.1365-313X.2011.04687.x URL |
| [16] |
Mao XG, Zhang HY, Qian XY, et al. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis[J]. J Exp Bot, 2012, 63(8): 2933-2946.
doi: 10.1093/jxb/err462 URL |
| [17] |
Xu ZY, Kim SY, Hyeon DY, et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses[J]. Plant Cell, 2013, 25(11): 4708-4724.
doi: 10.1105/tpc.113.119099 URL |
| [18] |
Yang SD, Seo PJ, Yoon HK, et al. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes[J]. Plant Cell, 2011, 23(6): 2155-2168.
doi: 10.1105/tpc.111.084913 URL |
| [19] |
Nakashima K, Tran LS P, Van Nguyen D, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice[J]. Plant J, 2007, 51(4): 617-630.
doi: 10.1111/j.1365-313X.2007.03168.x pmid: 17587305 |
| [20] |
Jeong JS, Kim YS, Baek KH, et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions[J]. Plant Physiol, 2010, 153(1): 185-197.
doi: 10.1104/pp.110.154773 pmid: 20335401 |
| [21] |
Bang SW, Choi S, Jin XJ, et al. Transcriptional activation of rice CINNAMOYL-CoA REDUCTASE 10 by OsNAC5, contributes to drought tolerance by modulating lignin accumulation in roots[J]. Plant Biotechnol J, 2022, 20(4): 736-747.
doi: 10.1111/pbi.v20.4 URL |
| [22] |
Song SY, Chen Y, Chen J, et al. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress[J]. Planta, 2011, 234(2): 331-345.
doi: 10.1007/s00425-011-1403-2 URL |
| [23] |
Takada S, Hibara K, Ishida T, et al. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation[J]. Development, 2001, 128(7): 1127-1135.
doi: 10.1242/dev.128.7.1127 pmid: 11245578 |
| [24] |
Sablowski RWM, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA[J]. Cell, 1998, 92(1): 93-103.
doi: 10.1016/s0092-8674(00)80902-2 pmid: 9489703 |
| [25] |
Breeze E, Harrison E, McHattie S, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation[J]. Plant Cell, 2011, 23(3): 873-894.
doi: 10.1105/tpc.111.083345 URL |
| [26] |
Kim YS, Kim SG, Park JE, et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis[J]. Plant Cell, 2006, 18(11): 3132-3144.
doi: 10.1105/tpc.106.043018 URL |
| [27] | 马雪祺, 阴艳红, 冯婧娴, 等. 植物NAC转录因子研究进展[J]. 植物生理学报, 2021, 57(12): 2225-2234. |
| Ma XQ, Yin YH, Feng JX, et al. Research progress of NAC transcription factors in plant[J]. Plant Physiol J, 2021, 57(12): 2225-2234. | |
| [28] |
Nuruzzaman M, Manimekalai R, Sharoni AM, et al. Genome-wide analysis of NAC transcription factor family in rice[J]. Gene, 2010, 465(1/2): 30-44.
doi: 10.1016/j.gene.2010.06.008 URL |
| [29] |
Shiriga K, Sharma R, Kumar K, et al. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize[J]. Meta Gene, 2014, 2: 407-417.
doi: 10.1016/j.mgene.2014.05.001 pmid: 25606426 |
| [30] |
Shang HH, Li W, Zou CS, et al. Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns[J]. J Integr Plant Biol, 2013, 55(7): 663-676.
doi: 10.1111/jipb.12085 URL |
| [31] |
Liu SX, Wang XL, Wang HW, et al. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L[J]. PLoS Genet, 2013, 9(9): e1003790.
doi: 10.1371/journal.pgen.1003790 URL |
| [32] |
Golldack D, Li C, Mohan H, et al. Tolerance to drought and salt stress in plants: Unraveling the signaling networks[J]. Front Plant Sci, 2014, 5: 151.
doi: 10.3389/fpls.2014.00151 pmid: 24795738 |
| [33] |
Tran LS P, Nakashima K, Sakuma Y, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter[J]. Plant Cell, 2004, 16(9): 2481-2498.
doi: 10.1105/tpc.104.022699 URL |
| [34] |
Mao XG, Chen SS, Li A, et al. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis[J]. PLoS One, 2014, 9(1): e84359.
doi: 10.1371/journal.pone.0084359 URL |
| [35] |
Lu M, Ying S, Zhang DF, et al. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis[J]. Plant Cell Rep, 2012, 31(9): 1701-1711.
doi: 10.1007/s00299-012-1284-2 URL |
| [36] | 熊江. 枳PtAOS1和PtNAC72基因响应干旱胁迫机理研究[D]. 长沙: 湖南农业大学, 2020. |
| Xiong J. Study on the mechanisms of PtAOS1 and PtNAC72 responding to drought stress in Poncirus trifoliata(l.)raf.[D]. Changsha: Hunan Agricultural University, 2020. | |
| [37] |
Wang LB, Liu LJ, Ma YL, et al. Transcriptome profilling analysis characterized the gene expression patterns responded to combined drought and heat stresses in soybean[J]. Comput Biol Chem, 2018, 77: 413-429.
doi: S1476-9271(18)30464-X pmid: 30476702 |
| [38] |
Xu CS, Xia C, Xia ZQ, et al. Physiological and transcriptomic responses of reproductive stage soybean to drought stress[J]. Plant Cell Rep, 2018, 37(12): 1611-1624.
doi: 10.1007/s00299-018-2332-3 pmid: 30099610 |
| [39] |
Zhang XY, Yang ZR, Li Z, et al. De novo transcriptome assembly and co-expression network analysis of Cynanchum thesioides: identification of genes involved in resistance to drought stress[J]. Gene, 2019, 710: 375-386.
doi: 10.1016/j.gene.2019.05.055 URL |
| [40] |
Gao TT, Zhang ZJ, Liu XM, et al. Physiological and transcriptome analyses of the effects of exogenous dopamine on drought tolerance in apple[J]. Plant Physiol Biochem, 2020, 148: 260-272.
doi: 10.1016/j.plaphy.2020.01.022 URL |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [5] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [6] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [7] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [8] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [9] | 张新博, 崔浩亮, 史佩华, 高锦春, 赵顺然, 陶晨雨. 低起始量的免疫共沉淀技术研究进展[J]. 生物技术通报, 2023, 39(4): 227-235. |
| [10] | 葛颜锐, 赵冉, 徐静, 李若凡, 胡云涛, 李瑞丽. 植物维管形成层发育及其调控的研究进展[J]. 生物技术通报, 2023, 39(3): 13-25. |
| [11] | 刘铖霞, 孙宗艳, 罗云波, 朱鸿亮, 曲桂芹. bHLH转录因子的磷酸化调控植物生理功能的研究进展[J]. 生物技术通报, 2023, 39(3): 26-34. |
| [12] | 赵孟良, 郭怡婷, 任延靖. 菊芋WRKY转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(2): 116-125. |
| [13] | 韩芳英, 胡昕, 王楠楠, 谢裕红, 王晓艳, 朱强. DREBs响应植物非生物逆境胁迫研究进展[J]. 生物技术通报, 2023, 39(11): 86-98. |
| [14] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [15] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||