








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 330-342.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1005
秦健1,2( ), 李振月1, 何浪1, 李俊玲1, 张昊1, 杜荣1(
), 李振月1, 何浪1, 李俊玲1, 张昊1, 杜荣1( )
)
收稿日期:2023-10-30
出版日期:2024-06-26
发布日期:2024-06-24
通讯作者:
杜荣,女,博士,教授,研究方向:动物细胞分子调控与生物工程;E-mail: drdurong@163.com作者简介:秦健,男,博士,副教授,研究方向:细胞分子调控与超微结构;E-mail: qinjian969@163.com
基金资助:
QIN Jian1,2( ), LI Zhen-yue1, HE Lang1, LI Jun-ling1, ZHANG Hao1, DU Rong1(
), LI Zhen-yue1, HE Lang1, LI Jun-ling1, ZHANG Hao1, DU Rong1( )
)
Received:2023-10-30
Published:2024-06-26
Online:2024-06-24
摘要:
【目的】 基于单细胞转录组测序(single-cell RNA sequencing, scRNA-seq)揭示牛肌源性细胞分化中的基因表达谱变化,并探究介导细胞间通讯的配体-受体互作机制,为构建成肌分化的动态调控网络奠定基础。【方法】 利用Seurat、ClusterProfiler、STRING、Cytoscape、CellChatDB和Monocle2等数据库或软件,对NCBI-GEO公共数据库中牛肌源性细胞单细胞转录组测序的原始数据进行了深入分析,包括细胞分群鉴定及差异基因表达谱、相关性、GO富集、PPI、细胞间通讯和拟时序分析等。【结果】 根据基因表达相关性及标志性基因共鉴定出4个具有独特转录特征的细胞群Myoblasts、Myocytes、Fibroblasts和FAPs,通过Myoblasts亚群的基因表达谱比较及拟时序分化轨迹分析发现,各亚群之间异质性很强,其中Myoblasts_1为分化轨迹起点,Myoblasts_0和3处于分化早期阶段,而Myoblasts_2是肌肉特征表现最为明显的亚群,可能是临近形成Myocytes的后期阶段的Myoblasts;Myoblasts_2和Myocytes差异基因富集的肌肉相关GO term存在差异,各基因间存在复杂的蛋白互作关系;Myoblasts_0-5、Myocytes、Fibroblasts和FAPs同型或异型细胞间的通讯机制,涉及到PTN-NCL、IGF2-IGF2R和ANGPTL2-(ITGA5 + ITGB1)等多种不同的配体-受体作用。【结论】 肌源性细胞在分化过程中存在不断变化的基因表达谱和细胞间通讯,反映了复杂的动态异质性及分子调控机制。
秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342.
QIN Jian, LI Zhen-yue, HE Lang, LI Jun-ling, ZHANG Hao, DU Rong. Change of Single-cell Transcription Profile and Analysis of Intercellular Communication in Myogenic Cell Differentiation[J]. Biotechnology Bulletin, 2024, 40(6): 330-342.
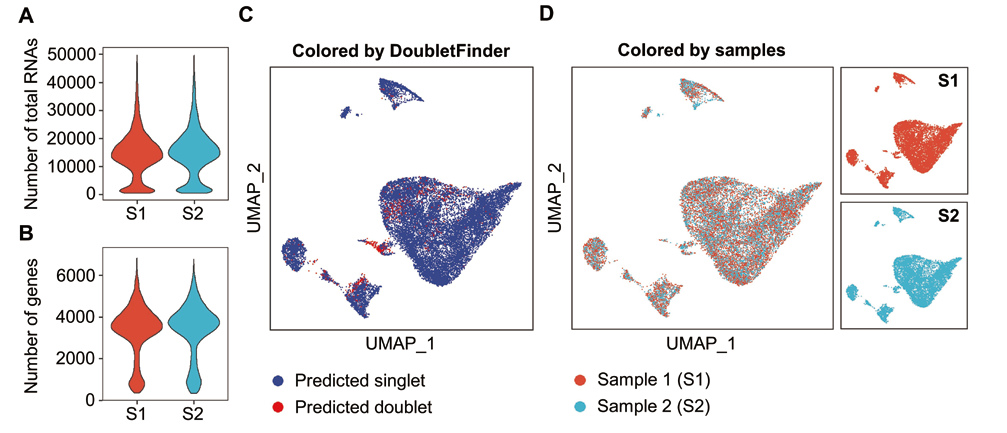
图1 牛肌源性细胞样本单细胞转录组测序数据的质量控制和样本整合分析 A: 小提琴图表示两个样本经过质量控制和过滤后剩余高质量细胞中的总RNA数量;B: 小提琴图表示两个样本经过质量控制和过滤后剩余高质量细胞中的总基因数量;C: UMAP降维图表示整合分析后两个样本的单细胞或双细胞性质预测结果(蓝色表示单细胞,红色表示双细胞);D: UMAP降维图表示对双细胞过滤后2个样本的单细胞展示(左图为合并展示,右图为分别展示。红色表示样本1,蓝色表示样本2)
Fig. 1 Quality control and sample integration analysis of scRNA-seq data from bovine myogenic cell samples A: A violin plot shows the number of total RNAs in the remaining high-quality cells of two samples after quality control and filtration. B: A violin plot shows the number of total genes in the remaining high-quality cells of two samples after quality control and filtration. C: A UMAP dimension reduction graph shows the singlet or doublet(single or double cells)property prediction results of two samples after integrated analysis(Blue indicates singlet and red indicates doublet). D: UMAP dimension reduction graphs show the singlet presentations of two samples after filtering the doublet(The left shows the combined display and the right shows the separate display. Red indicates sample 1 and blue indicates sample 2)

图2 单细胞转录组分析描绘的牛肌源性细胞样本中细胞类型图谱 A: UMAP降维图展示单细胞测序分析获得的12个细胞群(不同颜色表示不同的细胞群);B: 气泡图展示每个细胞群中标志性基因的表达水平(颜色标尺表示某基因在细胞群中的平均表达量,气泡大小表示表达某基因的细胞百分比);C: 热图展示12个细胞群之间基于差异基因表达水平的相关性(颜色标尺表示相关性系数);D: UMAP降维图展示根据标志基因和相关性分析将12个细胞群注释合并为4种细胞类型(不同颜色表示不同的细胞类型);E: UMAP叠加FeaturePlot展示每种细胞类型标志性基因的表达水平(颜色标尺表示基因表达量)
Fig. 2 Cell type profile of bovine myogenic cell samples delineated by scRNA-seq analysis A: A UMAP dimension reduction graph shows the 12 cell clusters obtained from scRNA-seq analysis(Different colors indicate the different cell clusters). B: A dot plot shows the expression levels of marker genes in each cell cluster(Color scale indicates the average expression level of a certain gene in the cell clusters, and dot size indicates the percentage of cells expressing a certain gene). C: A heatmap shows the correlations among 12 cell clusters based on the differential gene expression levels(Color scale indicates correlation coefficient). D: A UMAP dimension reduction graph shows the 12 cell clusters are annotated into 4 cell types based on the marker gene and correlation analysis(Different colors indicate the different cell types). E: UMAP graphs combined with FeaturePlots show the expression levels of marker genes for each cell type(Color scale indicates the gene expression level)

图3 牛Myoblasts的细胞亚群及其差异基因表达谱比较 A: 圈图展示两个牛肌源性细胞样本中4种细胞类型的比例;B: UMAP降维图展示两个样本合并后用于划分亚群的Myoblasts(蓝色表示Myoblasts);C: UMAP降维图展示分析获得的6个Myoblasts亚群(不同颜色表示不同的细胞亚群);D: 气泡图展示Myoblasts亚群代表性差异基因表达谱的比较(颜色标尺表示某基因在细胞亚群中的平均表达量,气泡大小表示表达某基因的细胞百分比)
Fig. 3 Cell subclusters and differential gene expression profile comparison of bovine Myoblasts A: Circle graphs show the proportions of four cell types from two samples of bovine myogenic cells. B: A UMAP dimension reduction graph shows the Myoblasts used to divide subclusters after the two samples are combined(Blue indicates Myoblasts). C: A UMAP dimension reduction graph shows the six Myoblasts subclusters(Different colors indicate the different cell subclusters). D: A dot plot shows the comparison of the representative differential gene expression profiles of the Myoblasts subclusters(Color scale indicates the average expression level of a certain gene in the cell subclusters, and dot size indicates the percentage of cells expressing a certain gene)
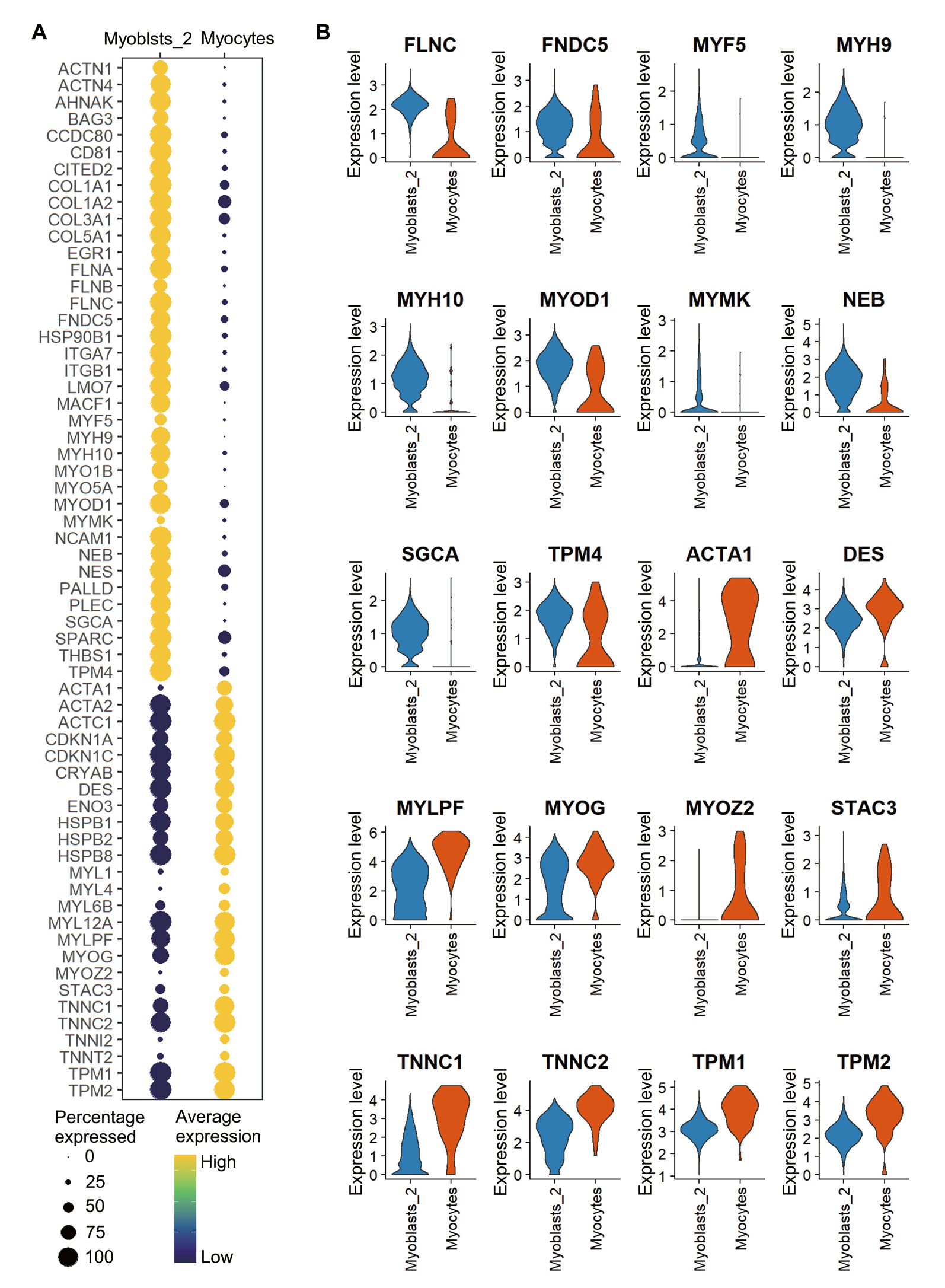
图4 Myoblasts_2与Myocytes之间主要肌肉相关差异基因的比较 A: 气泡图展示Myoblasts_2与Myocytes之间主要肌肉相关差异基因的表达谱(颜色标尺表示某基因在细胞群或亚群中的平均表达量,气泡大小表示表达某基因的细胞百分比);B: 小提琴图展示部分肌肉特异性差异基因在Myoblasts_2与Myocytes的表达水平和细胞分布情况(横坐标表示细胞类型,纵坐标表示基因表达水平,小提琴的宽度表示有对应表达水平的细胞密度,宽度越大说明该水平细胞占比越多)
Fig. 4 Comparison of the major muscle-related differential genes between Myoblasts_2 and Myocytes A: A dot plot shows the expression profile of the major muscle-related differential genes between Myoblasts_2 and Myocytes(Color scale indicates the average expression level of a certain gene in the cell cluster or subcluster, and dot size indicates the percentage of cells expressing a certain gene). B: Violin plots show the expression levels and cell distributions of some muscle-specific differential genes in Myoblasts_2 and Myocytes(Horizontal coordinate indicates the cell type and vertical coordinate indicates the gene expression level. The width of the violin indicates the cell density with the corresponding expression level, the larger the width, the more cells at the expression level)

图5 Myoblasts_2与Myocytes差异基因在肌肉相关生物过程的GO富集及PPI分析 A: Myoblasts_2中上调基因富集到的肌肉相关GO条目;B: Myocytes中上调基因富集到的肌肉相关GO条目;C: Myoblasts_2与Myocytes富集到肌肉相关GO条目中基因的PPI分析(黄色和绿色分别表示Myoblasts_2和Myocytes中上调基因)
Fig. 5 GO enrichment and PPI analysis of the Myoblasts_2 and Myocytes differential genes in muscle-related biological processes A: Muscle-related GO terms enriched by the up-regulated genes in Myoblasts_2. B: Muscle-related GO terms enriched by the up-regulated genes in Myocytes. C: PPI analysis of genes in muscle-related GO terms enriched by Myoblasts_2 and Myocytes(Yellow and green indicate the up-regulated genes respectively in Myoblasts_2 and Myocytes)

图6 牛肌源性细胞样本中各类型细胞间通讯的配体-受体分析 A: UMAP降维图展示6个Myoblasts亚群投射在细胞群中的位置关系(不同颜色表示不同的细胞群或亚群);B: 圆形网络图表示各细胞类型间的相互作用强度(周围圆形节点的大小表示某细胞类型中细胞的数量,颜色表示何种细胞为发送者,发出箭头的细胞为发送者,箭头指向的细胞为接收者,连线粗细表示互作强度);C: 气泡图展示细胞间通讯涉及的主要配体-受体(气泡大小表示P-Value,气泡颜色表示配体-受体的通讯概率);D: 小提琴图展示PTN配体和SDC/NCL受体基因在各细胞类型的表达水平和细胞分布情况(横坐标表示细胞类型,纵坐标表示基因表达水平,小提琴的宽度表示有对应表达水平的细胞密度,宽度越大说明该水平细胞占比越多)
Fig. 6 Analyses of ligand-receptor for intercellular communication of various cell types in bovine myogenic cell samples A: A UMAP dimension reduction graph shows the position relationship of the six myoblasts subclusters projected on the cell clusters(Different colors indicate the different cell clusters or subclusters). B: A circle plot shows the interaction strength between various cell types(The size of the surrounding circular nodes indicates the cell number in a certain cell type. The color indicates which cell is the sender, the cell that sends the arrow is the sender, and the cell that the arrow points to is the receiver. The thickness of the line indicates the interaction strength). C: A dot plot shows the major ligand-receptor pairs involved in intercellular communication(Dot size indicates the P-Value, and dot color indicates the ligand-receptor communication probability). D: A violin plot shows the expression levels and cell distributions of PTN ligand and SDC/NCL receptor genes in each cell type(Horizontal coordinate indicates the cell type and vertical coordinate indicates the gene expression level. The width of the violin indicates the cell density with the corresponding expression level, the larger the width, the more cells at the expression level)
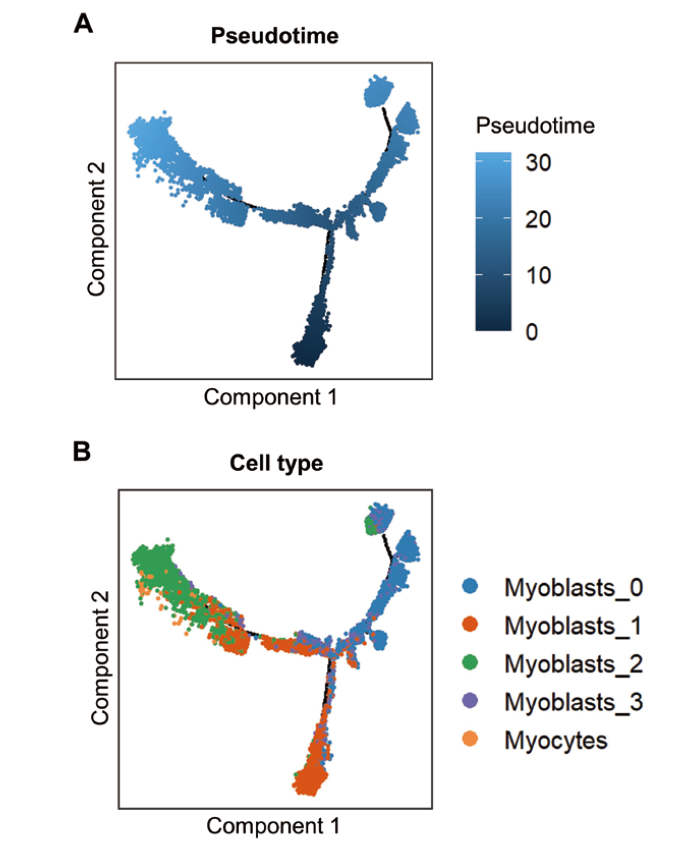
图7 基于拟时序分析构建的牛肌源性细胞亚群分化轨迹 A: 根据拟时序展示的细胞轨迹图(颜色由深至浅表示拟时序的起点至终点);B: 根据细胞类型展示的细胞轨迹图(不同颜色表示不同的细胞类型)
Fig. 7 Differentiation trajectory of bovine myogenic cell subclusters based on pseudotime analysis A: Cell trajectory plot displayed by pseudotime(The color from dark to light indicates the beginning to end of pseudotime). B: Cell trajectory plot displayed by cell type(Different colors indicate different cell types)
| [1] | Li JJ, Pei YL, Zhou R, et al. Regulation of RNA N6-methyladenosine modification and its emerging roles in skeletal muscle development[J]. Int J Biol Sci, 2021, 17(7): 1682-1692. |
| [2] | Rodríguez-Fdez S, Bustelo XR. Rho GTPases in skeletal muscle development and homeostasis[J]. Cells, 2021, 10(11): 2984. |
| [3] |
Weskamp K, Olwin BB, Parker R. Post-transcriptional regulation in skeletal muscle development, repair, and disease[J]. Trends Mol Med, 2021, 27(5): 469-481.
doi: 10.1016/j.molmed.2020.12.002 pmid: 33384234 |
| [4] | Cao JY, Spielmann M, Qiu XJ, et al. The single-cell transcriptional landscape of mammalian organogenesis[J]. Nature, 2019, 566(7745): 496-502. |
| [5] | Cai CC, Yue Y, Yue BL. Single-cell RNA sequencing in skeletal muscle developmental biology[J]. Biomed Pharmacother, 2023, 162: 114631. |
| [6] |
Tang FC, Barbacioru C, Wang YZ, et al. mRNA-Seq whole-transcriptome analysis of a single cell[J]. Nat Methods, 2009, 6(5): 377-382.
doi: 10.1038/nmeth.1315 pmid: 19349980 |
| [7] |
Zhang XN, Li TQ, Liu F, et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems[J]. Mol Cell, 2019, 73(1): 130-142.e5.
doi: S1097-2765(18)30880-3 pmid: 30472192 |
| [8] |
Xi HB, Langerman J, Sabri S, et al. A human skeletal muscle atlas identifies the trajectories of stem and progenitor cells across development and from human pluripotent stem cells[J]. Cell Stem Cell, 2020, 27(1): 158-176.e10.
doi: S1934-5909(20)30156-9 pmid: 32396864 |
| [9] | Cai SF, Hu B, Wang XY, et al. Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig[J]. BMC Biol, 2023, 21(1): 19. |
| [10] | McKellar DW, Walter LD, Song LT, et al. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration[J]. Commun Biol, 2021, 4(1): 1280. |
| [11] | Lyu PC, Qi YM, Tu ZJ, et al. Single-cell RNA sequencing reveals heterogeneity of cultured bovine satellite cells[J]. Front Genet, 2021, 12: 742077. |
| [12] | Cai CC, Wan P, Wang H, et al. Transcriptional and open chromatin analysis of bovine skeletal muscle development by single-cell sequencing[J]. Cell Prolif, 2023, 56(9): e13430. |
| [13] |
Wang LS, Gao PD, Li CY, et al. A single-cell atlas of bovine skeletal muscle reveals mechanisms regulating intramuscular adipogenesis and fibrogenesis[J]. J Cachexia Sarcopenia Muscle, 2023, 14(5): 2152-2167.
doi: 10.1002/jcsm.13292 pmid: 37439037 |
| [14] |
Satija R, Farrell JA, Gennert D, et al. Spatial reconstruction of single-cell gene expression data[J]. Nat Biotechnol, 2015, 33(5): 495-502.
doi: 10.1038/nbt.3192 pmid: 25867923 |
| [15] |
Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species[J]. Nat Biotechnol, 2018, 36(5): 411-420.
doi: 10.1038/nbt.4096 pmid: 29608179 |
| [16] |
Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data[J]. Cell, 2019, 177(7): 1888-1902.e21.
doi: S0092-8674(19)30559-8 pmid: 31178118 |
| [17] |
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors[J]. Cell Syst, 2019, 8(4): 329-337.e4.
doi: S2405-4712(19)30073-0 pmid: 30954475 |
| [18] |
Yu GC, Wang LG, Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287.
doi: 10.1089/omi.2011.0118 pmid: 22455463 |
| [19] | Jin SQ, Guerrero-Juarez CF, Zhang LH, et al. Inference and analysis of cell-cell communication using CellChat[J]. Nat Commun, 2021, 12(1): 1088. |
| [20] |
Qiu XJ, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories[J]. Nat Methods, 2017, 14(10): 979-982.
doi: 10.1038/nmeth.4402 pmid: 28825705 |
| [21] | Zhang XX, Lan YJ, Xu JY, et al. CellMarker: a manually curated resource of cell markers in human and mouse[J]. Nucleic Acids Res, 2019, 47(D1): D721-D728. |
| [22] | Hao DD, Becker N, Mückter E, et al. In vitro model of human skeletal muscle tissue for the study of resident macrophages and stem cells[J]. Biology, 2022, 11(6): 936. |
| [23] |
Chong JX, Talbot JC, Teets EM, et al. Mutations in MYLPF cause a novel segmental amyoplasia that manifests as distal arthrogryposis[J]. Am J Hum Genet, 2020, 107(2): 293-310.
doi: S0002-9297(20)30202-0 pmid: 32707087 |
| [24] |
Hall MN, Griffin CA, Simionescu A, et al. Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells[J]. Dev Biol, 2011, 357(1): 248-258.
doi: 10.1016/j.ydbio.2011.06.032 pmid: 21741962 |
| [25] |
Mancini M, Magnani E, Macchi F, et al. The multi-functionality of UHRF1: epigenome maintenance and preservation of genome integrity[J]. Nucleic Acids Res, 2021, 49(11): 6053-6068.
doi: 10.1093/nar/gkab293 pmid: 33939809 |
| [26] | Xiao FY, Jiang ZP, Yuan F, et al. Down-regulating NQO1 promotes cellular proliferation in K562 cells via elevating DNA synthesis[J]. Life Sci, 2020, 248: 117467. |
| [27] | Chen J, Chen LD, Hua J, et al. Long-term dynamic compression enhancement TGF-β3-induced chondrogenesis in bovine stem cells: a gene expression analysis[J]. BMC Genom Data, 2021, 22(1): 13. |
| [28] |
Wang Y, Chen L, Ju LG, et al. Novel biomarkers associated with progression and prognosis of bladder cancer identified by co-expression analysis[J]. Front Oncol, 2019, 9: 1030.
doi: 10.3389/fonc.2019.01030 pmid: 31681575 |
| [29] |
Calvo F, Ege N, Grande-Garcia A, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts[J]. Nat Cell Biol, 2013, 15(6): 637-646.
doi: 10.1038/ncb2756 pmid: 23708000 |
| [30] |
Fowler PC, Byrne DJ, O’Sullivan NC. Rare disease models provide insight into inherited forms of neurodegeneration[J]. J Rare Dis Res Treat, 2016, 1(3): 17-21.
pmid: 28603788 |
| [31] | Barbhuiya TK, Fisher M, Boittier ED, et al. Structural investigation of CDCA3-Cdh1 protein-protein interactions using in vitro studies and molecular dynamics simulation[J]. Protein Sci, 2023, 32(3): e4572. |
| [32] |
Fei F, Qu J, Li CY, et al. Role of metastasis-induced protein S100A4 in human non-tumor pathophysiologies[J]. Cell Biosci, 2017, 7: 64.
doi: 10.1186/s13578-017-0191-1 pmid: 29204268 |
| [33] |
Nigdelioglu R, Hamanaka RB, Meliton AY, et al. Transforming growth factor(TGF)-β promotes de novo serine synthesis for collagen production[J]. J Biol Chem, 2016, 291(53): 27239-27251.
doi: 10.1074/jbc.M116.756247 pmid: 27836973 |
| [34] | Cui C, Yin HD, Han SS, et al. Quantitative proteomic and phosphoproteomic analysis of chicken skeletal muscle during embryonic development[J]. Anim Biotechnol, 2023, 34(2): 122-133. |
| [35] | Daudon M, Ramé C, Estienne A, et al. Impact of fibronectin type III domain-containing family in the changes in metabolic and hormonal profiles during peripartum period in dairy cows[J]. Front Vet Sci, 2022, 9: 960778. |
| [36] | Sellathurai J, Nielsen J, Hejbøl EK, et al. Low oxygen tension enhances expression of myogenic genes when human myoblasts are activated from G0 arrest[J]. PLoS One, 2016, 11(7): e0158860. |
| [37] | Noureddine M, Gehmlich K. Structural and signaling proteins in the Z-disk and their role in cardiomyopathies[J]. Front Physiol, 2023, 14: 1143858. |
| [38] | Liu LY, Chen CL, Liu P, et al. MYH10 combines with MYH9 to recruit USP45 by deubiquitinating snail and promotes serous ovarian cancer carcinogenesis, progression, and cisplatin resistance[J]. Adv Sci, 2023, 10(14): e2203423. |
| [39] | Dube DK, Dube S, Abbott L, et al. Identification, characterization, and expression of sarcomeric tropomyosin isoforms in zebrafish[J]. Cytoskeleton, 2017, 74(3): 125-142. |
| [40] | Maggio S, Canonico B, Ceccaroli P, et al. Modulation of the circulating extracellular vesicles in response to different exercise regimens and study of their inflammatory effects[J]. Int J Mol Sci, 2023, 24(3): 3039. |
| [41] |
Paradžik M, Humphries JD, Stojanović N, et al. KANK2 links αVβ5 focal adhesions to microtubules and regulates sensitivity to microtubule poisons and cell migration[J]. Front Cell Dev Biol, 2020, 8: 125.
doi: 10.3389/fcell.2020.00125 pmid: 32195252 |
| [42] |
Lohmeier-Vogel EM, Heeley DH. Biochemical comparison of Tpm1.1(α)and Tpm2.2(β)tropomyosins from rabbit skeletal muscle[J]. Biochemistry, 2016, 55(9): 1418-1427.
doi: 10.1021/acs.biochem.5b01140 pmid: 26863527 |
| [43] |
Wang X. Pleiotrophin: activity and mechanism[J]. Adv Clin Chem, 2020, 98: 51-89.
doi: S0065-2423(20)30015-9 pmid: 32564788 |
| [44] | Poimenidi E, Theodoropoulou C, Koutsioumpa M, et al. Vascular endothelial growth factor A(VEGF-A)decreases expression and secretion of pleiotrophin in a VEGF receptor-independent manner[J]. Vascul Pharmacol, 2016, 80: 11-19. |
| [45] |
Erikson DW, Burghardt RC, Bayless KJ, et al. Secreted phosphoprotein 1(SPP1, osteopontin)binds to integrin alpha v beta 6 on porcine trophectoderm cells and integrin alpha v beta 3 on uterine luminal epithelial cells, and promotes trophectoderm cell adhesion and migration[J]. Biol Reprod, 2009, 81(5): 814-825.
doi: 10.1095/biolreprod.109.078600 pmid: 19571258 |
| [46] | Frank JW, Seo H, Burghardt RC, et al. ITGAV (alpha v integrins)bind SPP1(osteopontin)to support trophoblast cell adhesion[J]. Reproduction, 2017, 153(5): 695-706. |
| [47] | Torrente Y, Bella P, Tripodi L, et al. Role of insulin-like growth factor receptor 2 across muscle homeostasis: implications for treating muscular dystrophy[J]. Cells, 2020, 9(2): 441. |
| [48] | Chen WT, You WJ, Valencak TG, et al. Bidirectional roles of skeletal muscle fibro-adipogenic progenitors in homeostasis and disease[J]. Ageing Res Rev, 2022, 80: 101682. |
| [49] | Hanley CJ, Waise S, Ellis MJ, et al. Single-cell analysis reveals prognostic fibroblast subpopulations linked to molecular and immunological subtypes of lung cancer[J]. Nat Commun, 2023, 14(1): 387. |
| [50] | Nawaz A, Bilal M, Fujisaka S, et al. Depletion of CD206+ M2-like macrophages induces fibro-adipogenic progenitors activation and muscle regeneration[J]. Nat Commun, 2022, 13(1): 7058. |
| [51] | Riparini G, Simone JM, Sartorelli V. FACS-isolation and culture of fibro-adipogenic progenitors and muscle stem cells from unperturbed and injured mouse skeletal muscle[J]. J Vis Exp, 2022(184). DOI: 10.3791/6398 |
| [52] | Cameron A, Wakelin G, Gaulton N, et al. Identification of underexplored mesenchymal and vascular-related cell populations in human skeletal muscle[J]. Am J Physiol Cell Physiol, 2022, 323(6): C1586-C1600. |
| [53] | Kadomatsu T, Endo M, Miyata K, et al. Diverse roles of ANGPTL2 in physiology and pathophysiology[J]. Trends Endocrinol Metab, 2014, 25(5): 245-254. |
| [1] | 吴昊, 刘紫微, 郑颖, 戴雅文, 时权. 单细胞水平解析人牙龈间充质干细胞异质性[J]. 生物技术通报, 2023, 39(7): 325-332. |
| [2] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [3] | 赵艳坤, 刘慧敏, 孟璐, 王成, 王加启, 郑楠. 大肠埃希菌异质性耐药的研究进展[J]. 生物技术通报, 2022, 38(9): 59-71. |
| [4] | 熊和丽, 沙茜, 刘韶娜, 相德才, 张斌, 赵智勇. 单细胞转录组测序技术在动物上的应用研究[J]. 生物技术通报, 2022, 38(3): 226-233. |
| [5] | 寇佳怡, 王玉玲, 曾睿琳, 兰道亮. 单细胞转录组测序技术及在哺乳动物上的应用[J]. 生物技术通报, 2022, 38(11): 41-48. |
| [6] | 杨宏亮, 袁桢, 钱徐佳志, 徐大伟. 大麦Thionin-like基因家族基因表达谱分析[J]. 生物技术通报, 2022, 38(10): 140-147. |
| [7] | 郑青波, 叶娜, 张哓兰, 包鹏甲, 王福彬, 任稳稳, 廖月姣, 阎萍, 潘和平. 天祝白牦牛退行期毛囊细胞亚群鉴定以及特征基因生物信息学分析[J]. 生物技术通报, 2022, 38(10): 262-272. |
| [8] | 叶娜, 张晓兰, 包鹏甲, 王兴东, 阎萍, 潘和平. 单细胞测序技术及其在毛囊发育中的应用[J]. 生物技术通报, 2021, 37(10): 245-256. |
| [9] | 朱庆元, 李天晴. 单细胞转录组测序技术在心脏发育、疾病以及医学中的应用[J]. 生物技术通报, 2021, 37(1): 145-154. |
| [10] | 曹燕亭, 刘延峰, 李江华, 刘龙, 堵国成. 基于细胞亚群调控提升生物合成效率的研究进展[J]. 生物技术通报, 2020, 36(4): 19-25. |
| [11] | 邢亚欣, 黄火清, 苏小运. 一个来源于稻平脐蠕孢的α-阿拉伯呋喃糖苷酶[J]. 生物技术通报, 2020, 36(4): 84-92. |
| [12] | 王丹蕊, 沈文丽, 魏子艳, 王尚, 邓晔. 单细胞测序技术在微生物生态领域中的应用[J]. 生物技术通报, 2020, 36(10): 237-246. |
| [13] | 王娜, 龚娜, 刘国丽, 马晓颖, 杨镇, 杨涛. 内生菌次生代谢产物提高玉米渗透胁迫抗性的表达谱分析[J]. 生物技术通报, 2016, 32(7): 87-92. |
| [14] | 彭勇, 陈尚武, 马会勤. 黑果枸杞果实成熟发育过程表达谱差异分析[J]. 生物技术通报, 2016, 32(11): 144-151. |
| [15] | 肖尚,邓崇飞,柯军,鄢成伟,孙文正,杨彬. pH对重组CHO细胞生长、单抗表达及质量的影响[J]. 生物技术通报, 2015, 31(12): 256-261. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||