








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (4): 18-27.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1128
Previous Articles Next Articles
GAO Ling( ), WANG Fei, XIE Shuang-quan, CHEN Xi-feng, SHEN Hai-tao, LI Hong-bin(
), WANG Fei, XIE Shuang-quan, CHEN Xi-feng, SHEN Hai-tao, LI Hong-bin( )
)
Received:2020-09-05
Online:2021-04-26
Published:2021-05-13
Contact:
LI Hong-bin
E-mail:1425446422@qq.com;lihb@shzu.edu.cn
GAO Ling, WANG Fei, XIE Shuang-quan, CHEN Xi-feng, SHEN Hai-tao, LI Hong-bin. Genome-wide Identification and Expression Analysis of CBL Gene Family in Glycyrrhiza uralensis[J]. Biotechnology Bulletin, 2021, 37(4): 18-27.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物功能 Primer function |
|---|---|---|
| GuCBL1-F | CGAGCTTGACGTTGGGAGAG | GuCBL的RT-PCR检测 qRT-PCR detection of GuCBL |
| GuCBL1-R | CTGGCGAGGGCGACAATATC | |
| GuCBL2-F | TTTGTTTGCTGATCGGGTGT | |
| GuCBL2-R | ATTCCGCAAGCGTAGCAACT | |
| GuCBL3-F | CCGATTCGTCAAAGCAACCT | |
| GuCBL3-R | AGGATCCCATTGTGCTTTGTG | |
| GuCBL4-F | GAGAGGCGGTTTGCGTATTG | |
| GuCBL4-R | GCCTTTTTACGGTTCCTGGC | |
| GuCBL5-F | TGTCAGTGAGGTTGAGGCATT | |
| GuCBL5-R | CGCATTTGGGTGGAAGACAT | |
| GuCBL6-F | GCAGTGCAGTTATTGACGATGG | |
| GuCBL6-R | TGATGCACTGGGATGGAAGAC | |
| GuCBL7-F | GCTGTGTGCGGTGTTCATTC | |
| GuCBL7-R | GCGCAACGCCTCAATTTCAT | |
| GuCBL8-F | ATGAGGATCCCACCGTTCTTG | |
| GuCBL8-R | ACCCCATTGCGCTTGACATC | |
| GuCBL9-F | TCCTTGCTTCTGAAACACCCT | |
| GuCBL9-R | TGGCCATTGCGATTGACATC | |
| GuCBL10-F | CAGCAACAGCACAGGCATTT | |
| GuCBL10-R | CAGCAAGGCGAGCGAAATC | |
| GuActin-F | CCTCAACCCAAAGGTCAACAG | 内参基因 Internal reference gene |
| GuActin-R | GACCAGCGAGATCCAAACGAA | |
| GuCBL4-1 | GAAATTGAAGCATTATATGATCTGT | 全长基因克隆 Full-length gene cloning |
| GuCBL4-2 | GGAGCCTATGGAAAAACGCCAGCAA | |
| GuCBL7-1 | ACTTCGTAGCTAGCACTCCG | |
| GuCBL7-2 | GACTGCATGATGGCGGAAAA | |
| GuCBL10-1 | AGCAACAGCACAGGCATTTCC | |
| GuCBL10-2 | CAGCCTCCGATTTGAAAACAAAACT |
Table 1 Primers used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物功能 Primer function |
|---|---|---|
| GuCBL1-F | CGAGCTTGACGTTGGGAGAG | GuCBL的RT-PCR检测 qRT-PCR detection of GuCBL |
| GuCBL1-R | CTGGCGAGGGCGACAATATC | |
| GuCBL2-F | TTTGTTTGCTGATCGGGTGT | |
| GuCBL2-R | ATTCCGCAAGCGTAGCAACT | |
| GuCBL3-F | CCGATTCGTCAAAGCAACCT | |
| GuCBL3-R | AGGATCCCATTGTGCTTTGTG | |
| GuCBL4-F | GAGAGGCGGTTTGCGTATTG | |
| GuCBL4-R | GCCTTTTTACGGTTCCTGGC | |
| GuCBL5-F | TGTCAGTGAGGTTGAGGCATT | |
| GuCBL5-R | CGCATTTGGGTGGAAGACAT | |
| GuCBL6-F | GCAGTGCAGTTATTGACGATGG | |
| GuCBL6-R | TGATGCACTGGGATGGAAGAC | |
| GuCBL7-F | GCTGTGTGCGGTGTTCATTC | |
| GuCBL7-R | GCGCAACGCCTCAATTTCAT | |
| GuCBL8-F | ATGAGGATCCCACCGTTCTTG | |
| GuCBL8-R | ACCCCATTGCGCTTGACATC | |
| GuCBL9-F | TCCTTGCTTCTGAAACACCCT | |
| GuCBL9-R | TGGCCATTGCGATTGACATC | |
| GuCBL10-F | CAGCAACAGCACAGGCATTT | |
| GuCBL10-R | CAGCAAGGCGAGCGAAATC | |
| GuActin-F | CCTCAACCCAAAGGTCAACAG | 内参基因 Internal reference gene |
| GuActin-R | GACCAGCGAGATCCAAACGAA | |
| GuCBL4-1 | GAAATTGAAGCATTATATGATCTGT | 全长基因克隆 Full-length gene cloning |
| GuCBL4-2 | GGAGCCTATGGAAAAACGCCAGCAA | |
| GuCBL7-1 | ACTTCGTAGCTAGCACTCCG | |
| GuCBL7-2 | GACTGCATGATGGCGGAAAA | |
| GuCBL10-1 | AGCAACAGCACAGGCATTTCC | |
| GuCBL10-2 | CAGCCTCCGATTTGAAAACAAAACT |
| 基因名称 Gene name | 染色体位置 Chromosome location | 开放阅读框长度 Open reading frame sequence length/bp | 氨基酸数目Amino acid number/aa | 分子量Molecular weight/kD | 等电点Isoelectric point | N-端氨基酸 N-terminal amino acid | 棕榈酰化位点 Palmitoylation site | 豆蔻酰化位点 Myristoylation site | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| GuCBL1 | Scaffold00021(+):357697-361866 | 828 | 275 | 31.68 | 4.99 | MMIDMDFS | √ | × | 质膜 Plasma membrane |
| GuCBL2 | Scaffold00027(+):170105-174365 | 699 | 232 | 26.76 | 4.77 | MVKRDIML | √ | × | 质膜 Plasma membrane |
| GuCBL3 | Scaffold00030(-):499662-505101 | 750 | 249 | 28.71 | 4.93 | MPDSDSLF | √ | × | 质膜 Plasma membrane |
| GuCBL4 | Scaffold00036(-):366361-373219 | 1 008 | 331 | 37.63 | 9.31 | WLIFQKLK | √ | × | 质膜 Plasma membrane |
| GuCBL5 | Scaffold00067(+):227402-231449 | 642 | 213 | 24.36 | 4.67 | MGCFNSKA | √ | √ | 质膜 Plasma membrane |
| GuCBL6 | Scaffold00088(-):140793-143426 | 681 | 226 | 25.72 | 4.57 | MVQFLDEL | √ | × | 质膜 Plasma membrane |
| GuCBL7 | Scaffold00219(-):429-5651 | 861 | 364 | 41.46 | 5.33 | MDFVASTP | √ | × | 质膜 Plasma membrane |
| GuCBL8 | Scaffold00461(-):93781-104251 | 651 | 216 | 24.87 | 4.76 | MGCYCSTS | √ | √ | 质膜 Plasma membrane |
| GuCBL9 | Scaffold00669(-):36499-39615 | 717 | 238 | 27.41 | 5.38 | MGCRSSKV | √ | √ | 质膜 Plasma membrane |
| GuCBL10 | Scaffold01818(+):19453-23753 | 735 | 269 | 31.13 | 5.35 | MLQQQHRH | √ | × | 质膜 Plasma membrane |
Table 2 Characteristic analysis of G. uralensis CBL family genes
| 基因名称 Gene name | 染色体位置 Chromosome location | 开放阅读框长度 Open reading frame sequence length/bp | 氨基酸数目Amino acid number/aa | 分子量Molecular weight/kD | 等电点Isoelectric point | N-端氨基酸 N-terminal amino acid | 棕榈酰化位点 Palmitoylation site | 豆蔻酰化位点 Myristoylation site | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| GuCBL1 | Scaffold00021(+):357697-361866 | 828 | 275 | 31.68 | 4.99 | MMIDMDFS | √ | × | 质膜 Plasma membrane |
| GuCBL2 | Scaffold00027(+):170105-174365 | 699 | 232 | 26.76 | 4.77 | MVKRDIML | √ | × | 质膜 Plasma membrane |
| GuCBL3 | Scaffold00030(-):499662-505101 | 750 | 249 | 28.71 | 4.93 | MPDSDSLF | √ | × | 质膜 Plasma membrane |
| GuCBL4 | Scaffold00036(-):366361-373219 | 1 008 | 331 | 37.63 | 9.31 | WLIFQKLK | √ | × | 质膜 Plasma membrane |
| GuCBL5 | Scaffold00067(+):227402-231449 | 642 | 213 | 24.36 | 4.67 | MGCFNSKA | √ | √ | 质膜 Plasma membrane |
| GuCBL6 | Scaffold00088(-):140793-143426 | 681 | 226 | 25.72 | 4.57 | MVQFLDEL | √ | × | 质膜 Plasma membrane |
| GuCBL7 | Scaffold00219(-):429-5651 | 861 | 364 | 41.46 | 5.33 | MDFVASTP | √ | × | 质膜 Plasma membrane |
| GuCBL8 | Scaffold00461(-):93781-104251 | 651 | 216 | 24.87 | 4.76 | MGCYCSTS | √ | √ | 质膜 Plasma membrane |
| GuCBL9 | Scaffold00669(-):36499-39615 | 717 | 238 | 27.41 | 5.38 | MGCRSSKV | √ | √ | 质膜 Plasma membrane |
| GuCBL10 | Scaffold01818(+):19453-23753 | 735 | 269 | 31.13 | 5.35 | MLQQQHRH | √ | × | 质膜 Plasma membrane |

Fig. 1 Chromosomal scaffold distribution of GuCBL genes in G. uralensis GuCBL genes are located on 10 individual scaffolds. Pairs of segmentally duplicated genes are indicated by black lines. Chromosomal scaffolds are showed with different colored circles

Fig. 2 Phylogenetic relationship analysis of the CBL family The CBL proteins of G. uralensis, A. thaliana, and G. max were indicated in red, green, and blue, respectively
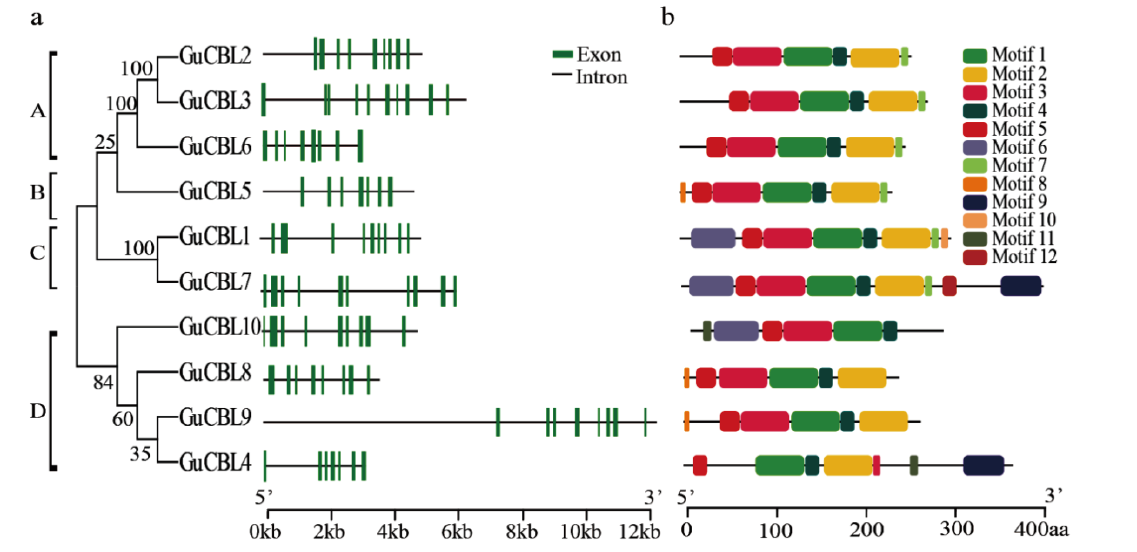
Fig. 3 Conserved motif and gene structure analyses of CBL family genes in G. uralensis a: Exon/intron structure analysis of GuCBL genes. Green boxes and black lines represent the exons and introns respectively, and the genomic length is indicated at the bottom. b: Conserved motif analysis of GuCBL proteins

Fig. 4 Multiple alignment and conserved motif analysis of CBL in G. uralensis EF-hand, PFPF-motif, myristoylation site, and serine residue of PFPF-motif are showed in red boxes

Fig. 5 Syntenic analysis of CBL genes in G. uralensis, G. max, and A. thaliana Blue, red, and purple lines represent duplicated CBL pairs in G. uralensis, syntenic relationships of CBL genes between G. uralensis and the other two species of G. max and A. thaliana, and CBL pairs between or inside G. max and A. thaliana
| 同源基因 Paralogous genes | Ka | Ks | Ka/Ks | 选择压 Selective pressure |
|---|---|---|---|---|
| GuCBL1-GuCBL7 | 0.2127 | 0.1996 | 1.0656 | Positive selection |
| GuCBL3-GuCBL2 | 0.1968 | 0.1674 | 1.1756 | Positive selection |
Table 3 Ka/Ks ratios for homologous CBL genes in G. uralensis
| 同源基因 Paralogous genes | Ka | Ks | Ka/Ks | 选择压 Selective pressure |
|---|---|---|---|---|
| GuCBL1-GuCBL7 | 0.2127 | 0.1996 | 1.0656 | Positive selection |
| GuCBL3-GuCBL2 | 0.1968 | 0.1674 | 1.1756 | Positive selection |
| 测试组 Testing group | 测试值Testing value | |||||
|---|---|---|---|---|---|---|
| Mtb | M1c | M2d | Χ2 | Pe | ||
| GuCBL1/GuCBL7 with GmCBL7 | 174 | 16 | 39 | 9.62 | 0.00193 | |
| GuCBL3/GuCBL2 with GmCBL6 | 236 | 19 | 5 | 8.27 | 0.00026 | |
Table 4 Tajima relative rate tests of homologous CBL gene pairs in G. uralensisa
| 测试组 Testing group | 测试值Testing value | |||||
|---|---|---|---|---|---|---|
| Mtb | M1c | M2d | Χ2 | Pe | ||
| GuCBL1/GuCBL7 with GmCBL7 | 174 | 16 | 39 | 9.62 | 0.00193 | |
| GuCBL3/GuCBL2 with GmCBL6 | 236 | 19 | 5 | 8.27 | 0.00026 | |
| [1] | 张继, 姚建, 丁兰, 等. 甘草的利用研究进展[J]. 草原与草坪, 2000,2:12-17. |
| Zhang J, Yao J, Ding L, et al. Study advances on the utilization of Glycyrrhiza[J]. Grassland and Turf, 2000,2:12-17. | |
| [2] | 廖建雄, 王根轩. 甘草酸在甘草适应荒漠生境中的可能作用[J]. 植物生理学通讯, 2003,39(4):367-370. |
| Liao JX, Wang GX. Possible role of glycyrrhizic acid in the adaptation of licorice to desert habitats[J]. Plant Physiology Communications, 2003,39(4):367-370. | |
| [3] | 万春阳. 盐分对甘草酸和甘草苷积累的影响及其作用机制研究[D]. 北京:北京中医药大学, 2011. |
| Wan CY. Effect of salt on the accumulation of glycyrrhizin and glycyrrhizin and the mechanism of action[D]. Beijing:Beijing University of Chinese Medicine, 2011. | |
| [4] | 卡迪尔·阿布都热西提. UV-B辐射与盐胁迫对两种甘草幼苗生长和生理生化特性的影响[D]. 西安:西北大学, 2018. |
| Kadir Abdulrashid. Effects of UV-B radiation and salt stress on the growth, physiological and biochemical characteristics of two licorice seedlings[D]. Xi’an:Northwestern University, 2018. | |
| [5] | 周雪洁. 干旱和外源激素对甘草生理特性及甘草酸积累的影响[D]. 杨凌:西北农林科技大学, 2011. |
| Zhou XJ. Effects of drought and exogenous hormones on physiological characteristics and glycyrrhizic acid accumulation of Licorice[D]. Yangling:Northwest A&F University, 2011. | |
| [6] | 梁晓薇. 茉莉酸甲酯对甘草次生代谢的调控[D]. 广州:广东药科大学, 2016. |
| Liang XW. Regulation of methyl jasmonate on secondary metabolism of Liquorice[D]. Guangzhou:Guangdong Pharmaceutical University, 2016. | |
| [7] | 林琳琳, 沈立, 林义章, 等. 钙信使在植物适应非生物胁迫中的作用[J]. 中国园艺文摘, 2015,31(4):41-45, 106. |
| Lin LL, Shen L, Lin YZ, et al. Role of calcium messenger in plant adaptation to abiotic stress[J]. Chinese Horticultural Digest, 2015,31(4):41-45, 106. | |
| [8] | 尚忠林, 毛国红, 孙大业. 植物细胞内钙信号的特异性[J]. 植物生理学通讯, 2003(2):93-100. |
| Shang ZL, Mao GH, Sun DY. Specificity of calcium signaling in plant cells[J]. Plant Physiology Communications, 2003(2):93-100. | |
| [9] |
Kolukisaoglu U, et al. Calcium sensors and their interacting protein kinases:genomics of the Arabidopsis and rice CBL-CIPK signaling networks[J]. Plant Physiology, 2004,134(1):43-58.
doi: 10.1104/pp.103.033068 URL |
| [10] |
Weinl S, Kudla J. The CBL-CIPK Ca2+-decoding signaling network:function and perspectives[J]. New Phytologist, 2009,184(3):517-528.
doi: 10.1111/nph.2009.184.issue-3 URL |
| [11] |
Cheong YH, Pandey GK, Grant JJ, et al. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis[J]. The Plant Journal, 2007,52(2):223-239.
doi: 10.1111/tpj.2007.52.issue-2 URL |
| [12] |
Albrecht V, Ritz O, Linder S, et al. The NAF domain defines a novel protein-protein interaction module in Ca2+-regulated kinases[J]. The EMBO Journal, 2001,20:1051-1063.
doi: 10.1093/emboj/20.5.1051 URL |
| [13] |
Pandey GK, et al. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynjournal in Arabidopsis[J]. The Plant Cell, 2004,16(7):1912-1924.
doi: 10.1105/tpc.021311 URL |
| [14] |
Nozawa A, Koizumi N, Sano H. An Arabidopsis SNF1-related protein kinase, AtSR1, interacts with a calcium-binding protein, AtCBL2, of which transcripts respond to light[J]. Plant and Cell Physiology, 2001,42(9):976-981.
pmid: 11577192 |
| [15] |
Xu J, Li HD, Chen LQ, et al. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis[J]. Cell, 2006,125(7):1347-1360.
doi: 10.1016/j.cell.2006.06.011 URL |
| [16] |
Wang M, Gu D, Liu T, et al. Overexpression of a putative maize calcineurin B -like protein in Arabidopsis confers salt tolerance[J]. Plant Molecular Biology, 2007,65(6):733-746.
doi: 10.1007/s11103-007-9238-8 URL |
| [17] | Gao P, Zhao PM, Wang J, et al. Co-expression and preferential interaction between two calcineurin B-like proteins and a CBL-interacting protein kinase from cotton[J]. Plant Physiology Biochemistry, 2008,46(10):925-940. |
| [18] | 马春英, 王文全, 赵玉新, 等. 乌拉尔甘草叶片解剖结构的研究[J]. 中国中药杂志, 2009,34(8):1034-1037. |
| Ma CY, Wang WQ, Zhao YX, et al. Study on the anatomical structure of leaves of Glycyrrhiza uralensis[J]. China Journal of Chinese Materia Medica, 2009,34(8):1034-1037. | |
| [19] | 罗江川. 乌拉尔甘草盐分积累、分布与分泌途径的研究[D]. 石河子:石河子大学, 2017. |
| Luo JC. Study on salt accumulation, distribution and secretory pathway of Glycyrrhiza uralensis[D]. Shihezi:Shihezi University, 2017. | |
| [20] | 彭伟秀, 王文全, 梁海永, 等. 水分胁迫对甘草营养器官解剖构造的影响[J]. 河北农业大学学报, 2003,26(3):46-48. |
| Peng WX, Wang WQ, Liang HY, et al. Effects of water stress on the anatomical structure of vegetative organs in licorice[J]. Journal of Hebei Agricultural University, 2003,26(3):46-48. | |
| [21] | Hall MA. 植物结构、功能和适应性[M]. 姚璧君译. 北京: 科学出版社, 1987. |
| Hall MA. Plant structure, function and adaptability[M]. Yao BJ, Trans. Beijing:Science Press, 1987. | |
| [22] | Rouffa AS, Carlquists S. Ecological strategies of xylem evolution[M]. Berkley University of California Press, 1975. |
| [23] | 陆嘉惠. 三种药用甘草耐盐性及耐盐机制研究[D]. 石河子:石河子大学, 2014. |
| Lu JH. Study on salt-tolerance and salt-tolerance mechanism of three medicinal Licorices[D]. Shihezi:Shihezi University, 2014. | |
| [24] | 谢亚军, 王兵, 等. 干旱胁迫对甘草幼苗活性氧代谢及保护酶活性的影响[J]. 农业科学研究, 2008,29(4):19-22. |
| Xie YJ, Wang B, et al. Effects of drought stress on active oxygen metabolism and protective enzyme activity in licorice seedlings[J]. Journal of Agricultural Sciences, 2008,29(4):19-22. | |
| [25] | 王芳. 甘草肌动蛋白基因GuActin2的克隆和表达分析[J]. 植物生理学通讯, 2009,45(10):995-1000. |
| Wang F. Molecular cloning and expression analysis of an actin gene GuActin2 from Chinese Licorice(Glycyrrhiza uralensis Fisch.)[J]. Plant Physiology Communications, 2009,45(10):995-1000. | |
| [26] | 黄聪琳. 拟南芥CBL和CIPK参与冷胁迫信号通路的研究[D]. 兰州:兰州大学, 2011. |
| Huang CL. CBL and CIPK involved cold stress signal pathway in Arabidopsis thaliana[D]. Lanzhou:Lanzhou University, 2011. | |
| [27] | 王娇娇, 韩胜芳, 等. 钙依赖蛋白激酶(CDPKs)介导植物信号转导的分子基础[J]. 草业学报, 2009,18(3):241-250. |
| Wang JJ, Han SF, et al. Molecular basis of signal transduction mediated by calciumdependent protein kinases(CDPKs)in plants[J]. Acta Prataculturae Sinica, 2009,18(3):241-250. | |
| [28] |
Mahs A, et al. The calcineurin B-like Ca2+-sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis[J]. Molecular Plant, 2013,6(4):1149-1162.
doi: 10.1093/mp/sst095 URL |
| [29] |
Kurusu T, Hamada J, Nokajima H, et al. Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells[J]. Plant Physiology, 2010,153(2):678-692.
doi: 10.1104/pp.109.151852 URL |
| [30] | 赵晋锋, 余爱丽, 王寒玉, 等. 非生物逆境胁迫下ZmCIPK10基因表达分析[J] 生物技术进展, 2011,1(2):130-134. |
| Zhao JF, Yu AL, Wang HY, et al. Expression analysis of ZmCIPK10 under abiotic stress[J]. Prog Biotech, 2011,1(2):130-134. | |
| [31] |
Quan R, Lin H, et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress[J]. The Plant Cell, 2007,19:1415-1431.
doi: 10.1105/tpc.106.042291 URL |
| [32] | 许园园, 蔺经, 李晓刚, 等. 梨CBL基因家族全基因组序列的鉴定及非生物胁迫下的表达分析[J]. 中国农业科学, 2015,48(4):735-747. |
| Xu YY, Lin J, et al. Identification of whole genome sequence of pear CBL gene family and expression analysis under abiotic stress[J]. Scientia Agricultura Sinica, 2015,48(4):735-747. | |
| [33] | 张和臣. 逆境条件下胡杨CBL-CIPK信号途径转导的分子机制研究[D]. 北京:北京林业大学, 2010. |
| Zhang HC. Molecular mechanism of signal pathway transduction in Populus euphratica under stress conditions[D]. Beijing:Beijing Forestry University, 2010. | |
| [34] |
Tong X, Cao A, Wang F, et al. Calcium-dependent protein kinase genes in Glycyrrhiza uralensis appear to be involved in promoting the biosynjournal of glycyrrhizic acid and flavonoids under salt stress[J]. Molecules, 2019,24(9):1837.
doi: 10.3390/molecules24091837 URL |
| [1] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [2] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [3] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [4] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [5] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [6] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [7] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [8] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [9] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [10] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [11] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [12] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [13] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [14] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [15] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||