








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 54-65.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1384
Previous Articles Next Articles
CHEN Chen( ), HUANG Zhi-yang, YU Hai-yan, YUAN Hai-bin, TIAN Huai-xiang(
), HUANG Zhi-yang, YU Hai-yan, YUAN Hai-bin, TIAN Huai-xiang( )
)
Received:2021-11-03
Online:2022-10-26
Published:2022-11-11
Contact:
TIAN Huai-xiang
E-mail:chenchen@sit.edu.cn;tianhx@sit.edu.cn
CHEN Chen, HUANG Zhi-yang, YU Hai-yan, YUAN Hai-bin, TIAN Huai-xiang. Research Technology and Progress in Transcriptional Regulation in Prokaryotes[J]. Biotechnology Bulletin, 2022, 38(10): 54-65.
| 研究技术 Method | 英文名称/缩写 Name/Abbreviation | 优缺点 Advantages/Disadvantages | 适用性 Applicability | 参考文献 Reference | |
|---|---|---|---|---|---|
| 体外方法 | 凝胶电泳迁移率实验 | EMSA | 方法简单、灵敏度高,放射性标记探针安全性低成本高,电泳运行条件下蛋白质-DNA复合体不稳定 | 适用于验证转录因子与假定DNA结合位点直接相互作用及结合位点突变对结合作用的影响 | [ |
| 等温滴定量热法 | ITC | 蛋白质无需固定化或修饰,样品消耗量少,可区分结合常数相近的配体相互作用及比较结构与结合作用的关系,对温度适应范围广但难以解释复杂系统中的相互作用 | 适用于成分简单的超高/超低亲和力相互作用系统及复杂的相互作用,可获得丰富的热力学信息 | [ | |
| DNase I footprinting技术 | DNase I footprinting | 分辨率高、可区分同一DNA片段多个不连续结合位点,但需要较多蛋白质才能产生清晰足迹,易产生超敏位点受到切割 | 适用于未纯化蛋白样品的检测,判断同一片段是否存在多个结合位点获得结合序列及比较各自亲和力 | [ | |
| 微量热泳动技术 | MST | 对相互作用的分子大小或质量无选择性,具有较好的适用性,对结合亲和力准确测定,可检测低至pM级别的结合亲和力,可在溶液环境中进行,无需固定分子避免结合假阳性 | 适用于结合亲和力弱,样品量小,样品所处环境复杂的情况 | [ | |
| 体内方法 | 染色质-免疫共沉淀技术 | ChIP | 接近细胞内真实情况,可研究转录因子对启动子结合的动态过程,但实验重复性不佳,获得良好实验结果对经验依赖性较高,对实验环境的要求严格 | 适用于确定转录因子修饰位置及低丰度转录因子结合分析 | [ |
| 细菌单杂交 | B1H | 细菌转化效率高构建文库质粒容量更大,无需复杂的仪器,转录因子需要能在大肠杆菌中表达,可能存在假阳性和假阴性的情况,结果需要进一步验证 | 适用于未纯化蛋白样品的检测,缺少相应仪器,用分子生物学手段进行筛选,转录因子要求能在大肠杆菌中表达,可发现新的转录因子 | [ | |
Table 1 Summary of research techniques on transcriptional regulation in prokaryotes
| 研究技术 Method | 英文名称/缩写 Name/Abbreviation | 优缺点 Advantages/Disadvantages | 适用性 Applicability | 参考文献 Reference | |
|---|---|---|---|---|---|
| 体外方法 | 凝胶电泳迁移率实验 | EMSA | 方法简单、灵敏度高,放射性标记探针安全性低成本高,电泳运行条件下蛋白质-DNA复合体不稳定 | 适用于验证转录因子与假定DNA结合位点直接相互作用及结合位点突变对结合作用的影响 | [ |
| 等温滴定量热法 | ITC | 蛋白质无需固定化或修饰,样品消耗量少,可区分结合常数相近的配体相互作用及比较结构与结合作用的关系,对温度适应范围广但难以解释复杂系统中的相互作用 | 适用于成分简单的超高/超低亲和力相互作用系统及复杂的相互作用,可获得丰富的热力学信息 | [ | |
| DNase I footprinting技术 | DNase I footprinting | 分辨率高、可区分同一DNA片段多个不连续结合位点,但需要较多蛋白质才能产生清晰足迹,易产生超敏位点受到切割 | 适用于未纯化蛋白样品的检测,判断同一片段是否存在多个结合位点获得结合序列及比较各自亲和力 | [ | |
| 微量热泳动技术 | MST | 对相互作用的分子大小或质量无选择性,具有较好的适用性,对结合亲和力准确测定,可检测低至pM级别的结合亲和力,可在溶液环境中进行,无需固定分子避免结合假阳性 | 适用于结合亲和力弱,样品量小,样品所处环境复杂的情况 | [ | |
| 体内方法 | 染色质-免疫共沉淀技术 | ChIP | 接近细胞内真实情况,可研究转录因子对启动子结合的动态过程,但实验重复性不佳,获得良好实验结果对经验依赖性较高,对实验环境的要求严格 | 适用于确定转录因子修饰位置及低丰度转录因子结合分析 | [ |
| 细菌单杂交 | B1H | 细菌转化效率高构建文库质粒容量更大,无需复杂的仪器,转录因子需要能在大肠杆菌中表达,可能存在假阳性和假阴性的情况,结果需要进一步验证 | 适用于未纯化蛋白样品的检测,缺少相应仪器,用分子生物学手段进行筛选,转录因子要求能在大肠杆菌中表达,可发现新的转录因子 | [ | |
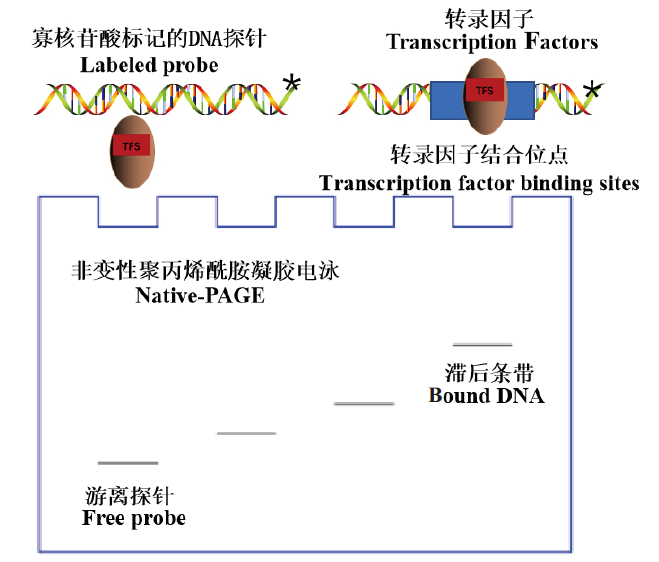
Fig. 1 Basic principle of EMSA experiment Asterisks indicate labeled probes,TFs indicate transcription factors,and TFBS indicate transcription factor binding sites
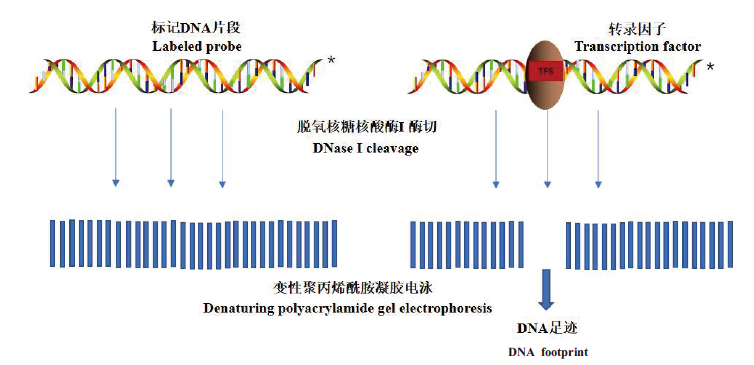
Fig. 2 Basic principle of DNase I footprint technology Adding appropriate concentration of deoxyribonuclease I(DNase I)to label strand in the DNA fragment,the DNA motif bound to the target protein will not be hydrolyzed by DNase I. After comparing the autoradiograph of the DNA fragment not bound to the target protein,interrupted DNA gradient bands can be obtained
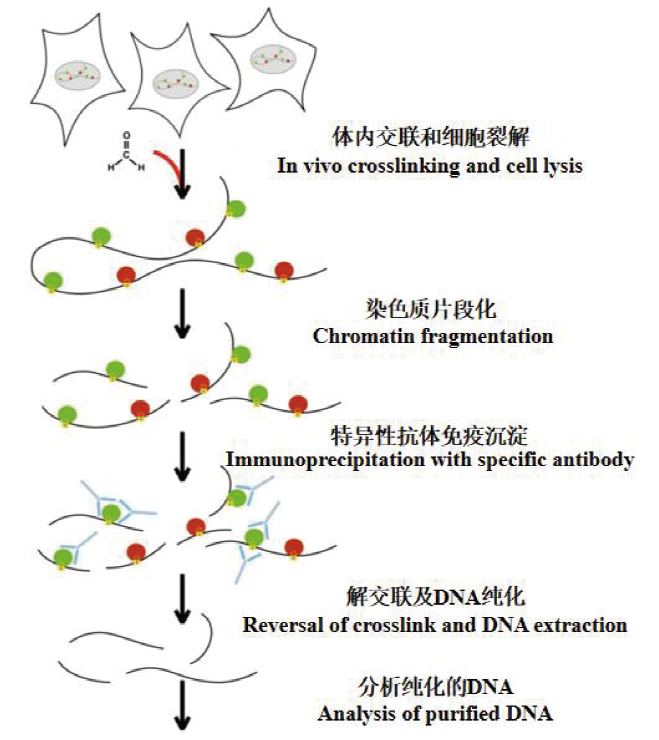
Fig. 3 Basic principle of ChIP experiment Cross-link the target protein with DNA in vivo through a cross-linking agent,and then fragment the DNA and protein complex. After that,precipitate with the specific antibody of the target protein to enrich the DNA fragments bound to the target protein. Reversal of crosslink and DNA extraction,finally analysis of purified DNA
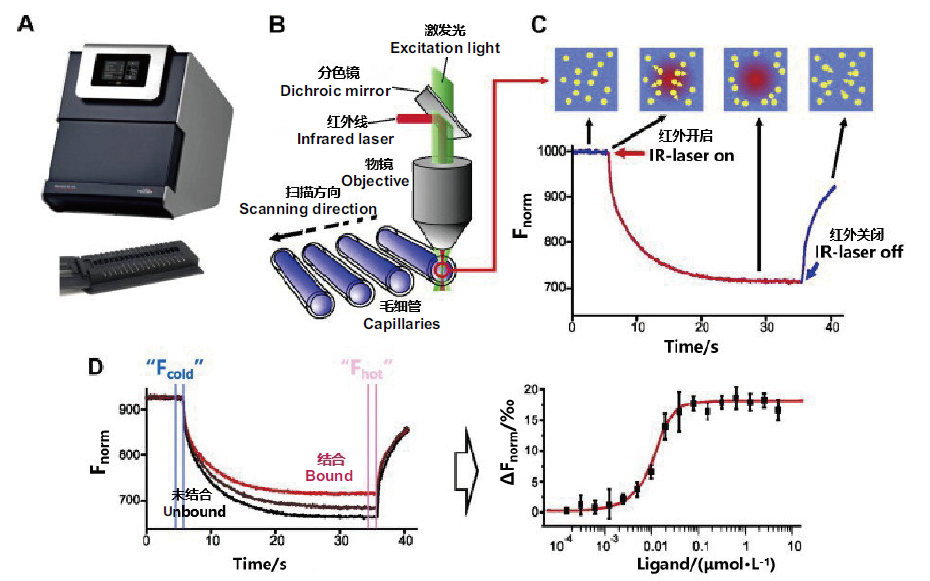
Fig. 4 Basic principle of MST experiment A:Fluorescence detector. B:MST optical element schematic diagram. C:MST experiment signal curve. D:MST combination experiment
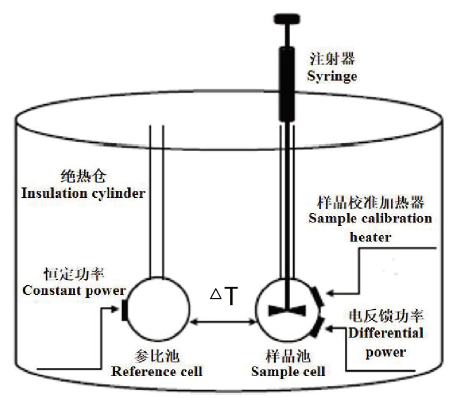
Fig. 5 Basic principle of ITC experiment The titration of reactants of known concentration into the sample causes the reaction between the components in the sample cell to be endothermic or exothermic. The temperature compensation system maintains a constant temperature difference between the sample cell and the reference cell,and the data are recorded to simulate the integrated heating isotherm. After that,information about the reaction enthalpy change,binding affinity and binding stoichiometry can be obtained.
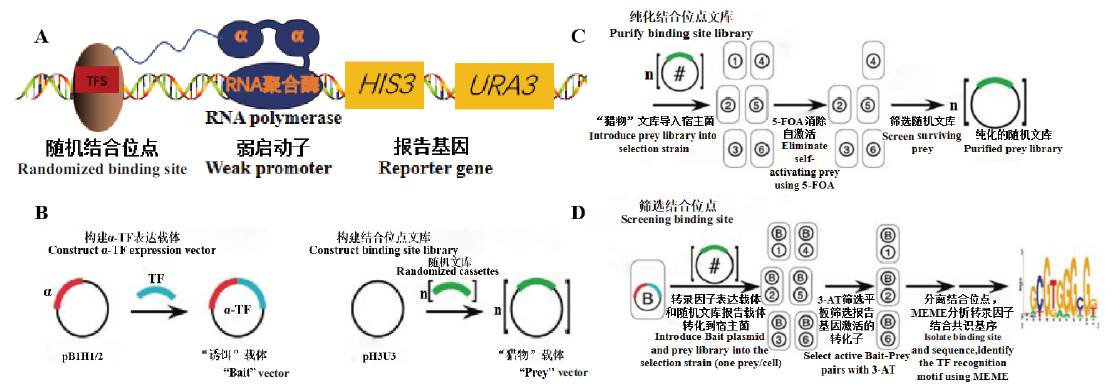
Fig. 6 Basic principle of bacterial one hybrid system A:Transcription factor recognition binding sites activate the expressions of the reporter genes. B:Construct α-TF expression vector and binding site library respectively. C:Eliminate self-activation to purify binding site library. D:Selection and analysis of binding sites
| [1] |
Fleischmann RD, Adams MD, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd[J]. Science, 1995, 269(5223):496-512.
pmid: 7542800 |
| [2] |
Faria JP, Overbeek R, Xia FF, et al. Genome-scale bacterial transcriptional regulatory networks:reconstruction and integrated analysis with metabolic models[J]. Brief Bioinform, 2014, 15(4):592-611.
pmid: 23422247 |
| [3] |
Kono N, Arakawa K. Nanopore sequencing:review of potential applications in functional genomics[J]. Dev Growth Differ, 2019, 61(5):316-326.
doi: 10.1111/dgd.12608 URL |
| [4] | 陈竺, 黄薇, 傅刚, 等. 人类基因组计划现状与展望[J]. 自然杂志, 2000, 22(3):125-133, 188. |
| Chen Z, Huang W, Fu G, et al. Human genome project progress and prospect[J]. Nat Mag, 2000, 22(3):125-133, 188. | |
| [5] |
Majewska M, Wysokińska H, Kuźma Ł, et al. Eukaryotic and prokaryotic promoter databases as valuable tools in exploring the regulation of gene transcription:a comprehensive overview[J]. Gene, 2018, 644:38-48.
doi: 10.1016/j.gene.2017.10.079 URL |
| [6] |
Monico C, Capitanio M, Belcastro G, et al. Optical methods to study protein-DNA interactions in vitro and in living cells at the single-molecule level[J]. Int J Mol Sci, 2013, 14(2):3961-3992.
doi: 10.3390/ijms14023961 URL |
| [7] | 周子康, 许平. 全局转录调控在细胞工厂构建中的应用与进展[J]. 化工进展, 2021, 40(3):1248-1251. |
| Zhou ZK, Xu P. Application and progress of global transcription regulation in microbial cell factory construction[J]. Chem Ind Eng Prog, 2021, 40(3):1248-1251. | |
| [8] |
Hellman LM, Fried MG. Electrophoretic mobility shift assay(EMSA)for detecting protein-nucleic acid interactions[J]. Nat Protoc, 2007, 2(8):1849-1861.
pmid: 17703195 |
| [9] |
Unterholzner SJ, Rozhon W, Poppenberger B. Analysis of in vitro DNA interactions of brassinosteroid-controlled transcription factors using electrophoretic mobility shift assay[J]. Methods Mol Biol, 2017, 1564:133-144.
doi: 10.1007/978-1-4939-6813-8_11 pmid: 28124251 |
| [10] |
齐心洁, 王玥, 等. 等温滴定量热法在蛋白质-配体相互作用中的应用[J]. 生物技术通报, 2017, 33(5):40-49.
doi: 10.13560/j.cnki.biotech.bull.1985.2017.05.006 |
| Qi XJ, Wang Y, et al. Applications of isothermal titration calorimetry in protein-ligand interactions[J]. Biotechnol Bull, 2017, 33(5):40-49. | |
| [11] |
Velazquez-Campoy A, Freire E. Isothermal titration calorimetry to determine association constants for high-affinity ligands[J]. Nat Protoc, 2006, 1(1):186-191.
pmid: 17406231 |
| [12] |
Velazquez-Campoy A, Freire E. ITC in the post-genomic era... ? priceless[J]. Biophys Chem, 2005, 115(2-3):115-124.
doi: 10.1016/j.bpc.2004.12.015 URL |
| [13] | Leblanc B, Moss T. DNase I footprinting[M]// Tom Moss. DNA-Protein Interactions. New Jersey: Humana Press, 2001:31-38. |
| [14] | 李丽琴, 石童, 周国超, 等. MST技术在生命科学中的应用进展[J]. 现代生物医学进展, 2017, 17(32):6393-6397. |
| Li LQ, Shi T, Zhou GC, et al. Progress on the applications of microscale themophoresis technology in life science[J]. Prog Mod Biomed, 2017, 17(32):6393-6397. | |
| [15] |
Gade P, Kalvakolanu DV. Chromatin immunoprecipitation assay as a tool for analyzing transcription factor activity[J]. Methods Mol Biol, 2012, 809:85-104.
doi: 10.1007/978-1-61779-376-9_6 pmid: 22113270 |
| [16] |
Christensen RG, Gupta A, Zuo Z, et al. A modified bacterial one-hybrid system yields improved quantitative models of transcription factor specificity[J]. Nucleic Acids Res, 2011, 39(12):e83.
doi: 10.1093/nar/gkr239 URL |
| [17] |
Bak G, Han K, et al. Electrophoretic mobility shift assay of RNA-RNA complexes[J]. Methods Mol Biol, 2015, 1240:153-163.
doi: 10.1007/978-1-4939-1896-6_12 pmid: 25352144 |
| [18] |
Künne T, Westra ER, Brouns SJJ. Electrophoretic mobility shift assay of DNA and CRISPR-cas ribonucleoprotein complexes[J]. Methods Mol Biol, 2015, 1311:171-184.
doi: 10.1007/978-1-4939-2687-9_11 pmid: 25981473 |
| [19] |
Pan YC, Karns K, Herr AE. Microfluidic electrophoretic mobility shift assays for quantitative biochemical analysis[J]. Electrophoresis, 2014, 35(15):2078-2090.
doi: 10.1002/elps.201300500 pmid: 24591076 |
| [20] |
Vavrova A, Vrzal R, Dvorak Z. A nonradioactive electrophoretic mobility shift assay for measurement of pregnane X receptor binding activity to CYP3A4 response element[J]. ELECTROPHORESIS, 2013, 34(13):1863-1668.
pmid: 23977680 |
| [21] |
Cornelussen RNM, Gupta S, Knowlton AA. Regulation of prostaglandin A1-induced heat shock protein expression in isolated cardiomyocytes[J]. J Mol Cell Cardiol, 2001(8):1447-1454.
pmid: 11448133 |
| [22] | 葛兴枫, 李慧, 李少华, 等. 用改良的非同位素凝胶电泳迁移实验鉴定核酸适配体与靶蛋白的结合[J]. 生物技术通讯, 2015, 26(2):249-251. |
| Ge XF, Li H, Li SH, et al. A modified non-isotope EMSA technique for identification of aptamer binding to its target protein[J]. Lett Biotechnol, 2015, 26(2):249-251. | |
| [23] |
Tokunaga S, Stegeman JJ. Elimination of nonspecific bands in non-radioactive electrophoretic mobility shift assays using the digoxigenin system[J]. Anal Biochem, 2014, 465:70-72.
doi: 10.1016/j.ab.2014.06.020 pmid: 25004462 |
| [24] |
Fried MG, Bromberg JL. Factors that affect the stability of protein-DNA complexes during gel electrophoresis[J]. Electrophoresis, 1997, 18(1):6-11.
pmid: 9059813 |
| [25] |
Brown L, Villegas JM, Elean M, et al. YebC, a putative transcriptional factor involved in the regulation of the proteolytic system of Lactobacillus[J]. Sci Rep, 2017, 7(1):8579.
doi: 10.1038/s41598-017-09124-1 URL |
| [26] | 王丽滨. 肺炎链球菌糖代谢蛋白CcpA对荚膜多糖的调控研究[D]. 重庆: 重庆医科大学, 2015. |
| Wang LB. The regulation effect of CcpA protein on the biosynthesis of capsular polysaccharide in Streptococcus pneumoniae[D]. Chongqing: Chongqing Medical University, 2015. | |
| [27] |
Chen C, Lu YQ, Wang LL, et al. CcpA-dependent carbon catabolite repression regulates fructooligosaccharides metabolism in Lactobacillus plantarum[J]. Front Microbiol, 2018, 9:1114.
doi: 10.3389/fmicb.2018.01114 pmid: 29896178 |
| [28] |
Keyhani J, Keyhani E. Detection of DNA autoantibodies by electrophoretic mobility shift assay[J]. Methods Mol Biol, 2019, 1901:133-152.
doi: 10.1007/978-1-4939-8949-2_11 pmid: 30539574 |
| [29] |
Hampshire AJ, Rusling DA, Broughton-Head VJ, et al. Footprinting:a method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands[J]. Methods, 2007, 42(2):128-140.
pmid: 17472895 |
| [30] | 徐冬冬, 刘德培, 吕湘, 等. 固相DNaseⅠ足迹法研究DNA-蛋白质相互作用[J]. 生物化学与生物物理进展, 2001, 28(4):587-590. |
| Xu DD, Liu DP, Lv X, et al. A method for the study of DNA-protein interaction:solid-phase DNaseⅠ footprinting[J]. Prog Biochem Biophys, 2001, 28(4):587-590. | |
| [31] | Gong LC, Ren C, Xu Y. GlnR negatively regulates glutamate-dependent acid resistance in Lactobacillus brevis[J]. Appl Environ Microbiol, 2020, 86(7):e02615-e02619. |
| [32] |
Ihara K, Sato K, Hori H, et al. Expression of the alaE gene is positively regulated by the global regulator Lrp in response to intracellular accumulation of l-alanine in Escherichia coli[J]. J Biosci Bioeng, 2017, 123(4):444-450.
doi: 10.1016/j.jbiosc.2016.11.015 URL |
| [33] | Yang XP, Teng KL, Li LL, et al. Transcriptional regulator AcrR increases ethanol tolerance through regulation of fatty acid synthesis in Lactobacillus plantarum[J]. Appl Environ Microbiol, 2019, 85(22):e01690-e01619. |
| [34] | 王智. DNase1足纹法[J]. 基础医学与临床, 1990, 10(3):59-63. |
| Wang Z. DNase I-footprinting assay[J]. Basic Med Sci Clin, 1990, 10(3):59-63. | |
| [35] |
Wagner M, Jung J, Koslowski M, et al. Chromatin immunoprecipitation assay to identify genomic binding sites of regulatory factors[J]. Methods Mol Biol, 2016, 1366:53-65.
doi: 10.1007/978-1-4939-3127-9_6 pmid: 26585127 |
| [36] |
Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin[J]. Cell, 1993, 75(6):1187-1198.
pmid: 7903220 |
| [37] | Wiehle L, Breiling A. Chromatin immunoprecipitation[M]// Lanzuolo C, Bodega B. Polycomb Group Proteins. New York: Humana Press, 2016:7-21. |
| [38] |
Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells[J]. Stem Cells, 2007, 25(4):1037-1046.
pmid: 17272500 |
| [39] |
Walton CB, Matter ML. Chromatin immunoprecipitation assay:examining the interaction of NFkB with the VEGF promoter[J]. Methods Mol Biol, 2015, 1332:75-87.
doi: 10.1007/978-1-4939-2917-7_6 pmid: 26285747 |
| [40] |
Chen H, Lin RJ, Xie W, et al. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase[J]. Cell, 1999, 98(5):675-686.
pmid: 10490106 |
| [41] |
Ishihama A. Prokaryotic genome regulation:multifactor promoters, multitarget regulators and hierarchic networks[J]. FEMS Microbiol Rev, 2010, 34(5):628-645.
doi: 10.1111/j.1574-6976.2010.00227.x pmid: 20491932 |
| [42] |
Ratib NR, Sabio EY, Mendoza C, et al. Genome-wide identification of genes directly regulated by ChvI and a consensus sequence for ChvI binding in Sinorhizobium meliloti[J]. Mol Microbiol, 2018, 110(4):596-615.
doi: 10.1111/mmi.14119 URL |
| [43] |
Yun CS, Takahashi Y, Shintani M, et al. MvaT family proteins encoded on IncP-7 plasmid pCAR1 and the host chromosome regulate the host transcriptome cooperatively but differently[J]. Appl Environ Microbiol, 2015, 82(3):832-842.
doi: 10.1128/AEM.03071-15 URL |
| [44] | Shao XL, Zhang XN, Zhang YC, et al. RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1[J]. J Bacteriol, 2018, 200(16):e00205-e00218. |
| [45] |
Bard-Chapeau EA, Jeyakani J, Kok CH, et al. Ecotopic viral integration site 1(EVI1)regulates multiple cellular processes important for cancer and is a synergistic partner for FOS protein in invasive tumors[J]. PNAS, 2012, 109(6):2168-2173.
doi: 10.1073/pnas.1119229109 pmid: 22308434 |
| [46] |
王泓力, 焦雨铃. 染色质免疫共沉淀实验方法[J]. 植物学报, 2020, 55(4):475-480.
doi: 10.11983/CBB20076 |
| Wang HL, Jiao YL. Protocols for chromatin immunoprecipitation[J]. Chin Bull Bot, 2020, 55(4):475-480. | |
| [47] |
Breitsprecher D, Schlinck N, Witte D, et al. Aptamer binding studies using MicroScale thermophoresis[J]. Methods Mol Biol, 2016, 1380:99-111.
doi: 10.1007/978-1-4939-3197-2_8 pmid: 26552819 |
| [48] |
Jerabek-Willemsen M, André T, Wanner R, et al. MicroScale thermophoresis:interaction analysis and beyond[J]. J Mol Struct, 2014, 1077:101-113.
doi: 10.1016/j.molstruc.2014.03.009 URL |
| [49] |
Jerabek-Willemsen M, Wienken CJ, Braun D, et al. Molecular interaction studies using microscale thermophoresis[J]. Assay Drug Dev Technol, 2011, 9(4):342-353.
doi: 10.1089/adt.2011.0380 URL |
| [50] | Gudim I, Lofstad M, Hammerstad M, et al. Measurement of FNR-NrdI interaction by microscale thermophoresis(MST)[J]. Bio-protocol, 2017, 7(8): e2223. |
| [51] |
Entzian C, Schubert T. Studying small molecule-aptamer interactions using microScale thermophoresis(MST)[J]. Methods, 2016, 97:27-34.
doi: 10.1016/j.ymeth.2015.08.023 URL |
| [52] |
Wienken CJ, Baaske P, Duhr S, et al. Thermophoretic melting curves quantify the conformation and stability of RNA and DNA[J]. Nucleic Acids Res, 2011, 39(8):e52.
doi: 10.1093/nar/gkr035 URL |
| [53] |
Zillner K, Filarsky M, Rachow K, et al. Large-scale organization of ribosomal DNA chromatin is regulated by Tip5[J]. Nucleic Acids Res, 2013, 41(10):5251-5262.
doi: 10.1093/nar/gkt218 pmid: 23580549 |
| [54] |
Shang X, Marchioni F, Evelyn CR, et al. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors[J]. PNAS, 2013, 110(8):3155-3160.
doi: 10.1073/pnas.1212324110 pmid: 23382194 |
| [55] |
van den Bogaart G, Meyenberg K, et al. Phosphatidylinositol 4, 5-bisphosphate increases Ca2+ affinity of synaptotagmin-1 by 40-fold[J]. J Biol Chem, 2012, 287(20):16447-16453.
doi: 10.1074/jbc.M112.343418 pmid: 22447935 |
| [56] |
Papageorgiou AC, Adam PS, et al. HU histone-like DNA-binding protein from Thermus thermophilus:structural and evolutionary analyses[J]. Extremophiles, 2016, 20(5):695-709.
doi: 10.1007/s00792-016-0859-1 pmid: 27342116 |
| [57] |
Seidel SAI, Dijkman PM, Lea WA, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions[J]. Methods, 2013, 59(3):301-315.
doi: 10.1016/j.ymeth.2012.12.005 pmid: 23270813 |
| [58] | 吴萌, 李竑, 陈铭. 两种实验技术在蛋白质-蛋白质相互作用检测中的应用[J]. 生命的化学, 2021(2):353-360. |
| Wu M, Li H, Chen M. An overview of BLI and MST applications in protein-protein interaction[J]. Chem Life, 2021(2):353-360. | |
| [59] |
Wang Q, Wang J, Song SX, et al. Microscale thermophoresis in the investigation of biomolecular interactions[J]. J Chin Pharm Sci, 2020, 29(9):656-665.
doi: 10.5246/jcps.2020.09.061 |
| [60] |
Keller S, Vargas C, Zhao HY, et al. High-precision isothermal titration calorimetry with automated peak-shape analysis[J]. Anal Chem, 2012, 84(11):5066-5073.
doi: 10.1021/ac3007522 pmid: 22530732 |
| [61] |
Boudker O, Oh S. Isothermal titration calorimetry of ion-coupled membrane transporters[J]. Methods, 2015, 76:171-182.
doi: S1046-2023(15)00024-9 pmid: 25676707 |
| [62] |
Zhuo L, Zhang Z, Pan Z, et al. CIRCE element evolved for the coordinated transcriptional regulation of bacterial duplicate groELs[J]. Biochim Biophys Acta Gene Regul Mech, 2018, 1861(10):928-937.
doi: 10.1016/j.bbagrm.2018.08.003 URL |
| [63] |
Wang W, Ji J, Li X, et al. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor[J]. PNAS, 2014, 111(15):5688-5693.
doi: 10.1073/pnas.1324253111 URL |
| [64] |
Yamasaki K, Akutsu Y, Yamasaki T, et al. Enhanced affinity of racemic phosphorothioate DNA with transcription factor SATB1 arising from diastereomer-specific hydrogen bonds and hydrophobic contacts[J]. Nucleic Acids Res, 2020, 48(8):4551-4561.
doi: 10.1093/nar/gkaa170 pmid: 32187371 |
| [65] |
Weber A, Dettling M, Brunner H, et al. Isothermal titration calorimetry of molecularly imprinted polymer nanospheres[J]. Macromol Rapid Commun, 2002, 23(14):824-828.
doi: 10.1002/1521-3927(20021001)23:14<824::AID-MARC824>3.0.CO;2-P URL |
| [66] |
Brautigam CA, Zhao HY, et al. Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions[J]. Nat Protoc, 2016(5):882-894.
doi: 10.1038/nprot.2016.044 pmid: 27055097 |
| [67] |
Meng XD, Wolfe SA. Identifying DNA sequences recognized by a transcription factor using a bacterial one-hybrid system[J]. Nat Protoc, 2006, 1(1):30-45.
pmid: 17406209 |
| [68] | 翟征远. 德氏乳杆菌保加利亚亚种CAUH1酸耐受机制的蛋白组学研究及抗酸胁迫基因Ldb0677和pyk的功能分析[D]. 北京: 中国农业大学, 2014. |
| Zhai ZY. Proteomic characterization of the acid tolerance response in Lactobacillus delbrueckii subsp. bulgaricus CAUH1and functional identification of acid stress-related genes Ldb0677 and pyk[D]. Beijing: China Agricultural University, 2014. | |
| [69] | Hebdon SD, Menon SK, Richter-Addo GB, et al. Regulatory targets of the response regulator RR_1586 from Clostridioides difficile identified using a bacterial one-hybrid screen[J]. J Bacteriol, 2018, 200(23):e00351-e00318. |
| [70] |
Zhai ZY, Douillard FP, An HR, et al. Proteomic characterization of the acid tolerance response in Lactobacillus delbrueckii subsp. bulgaricus CAUH1 and functional identification of a novel acid stress-related transcriptional regulator Ldb0677[J]. Environ Microbiol, 2014, 16(6):1524-1537.
doi: 10.1111/1462-2920.12280 URL |
| [71] | Zhu LJ, Christensen RG, Kazemian M, et al. FlyFactorSurvey:a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system[J]. Nucleic Acids Res, 2011, 39(Database issue):D111-D117. |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [3] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [4] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [5] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [6] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [7] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [8] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [9] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [10] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [11] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [12] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [13] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [14] | XIONG Shu-qi. Towards the Understanding on the Physiological Functions of Bile Acids and Interactions with Gut Microbiota [J]. Biotechnology Bulletin, 2023, 39(4): 187-200. |
| [15] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||