








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (4): 176-186.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1001
Previous Articles Next Articles
AN Lei1,3( ), ZHAO Jin-ling1,3, REN Xiao-liang1,2(
), ZHAO Jin-ling1,3, REN Xiao-liang1,2( )
)
Received:2022-08-15
Online:2023-04-26
Published:2023-05-16
AN Lei, ZHAO Jin-ling, REN Xiao-liang. RNA Modification and Its Research Progress in Caenorhabditis elegans[J]. Biotechnology Bulletin, 2023, 39(4): 176-186.
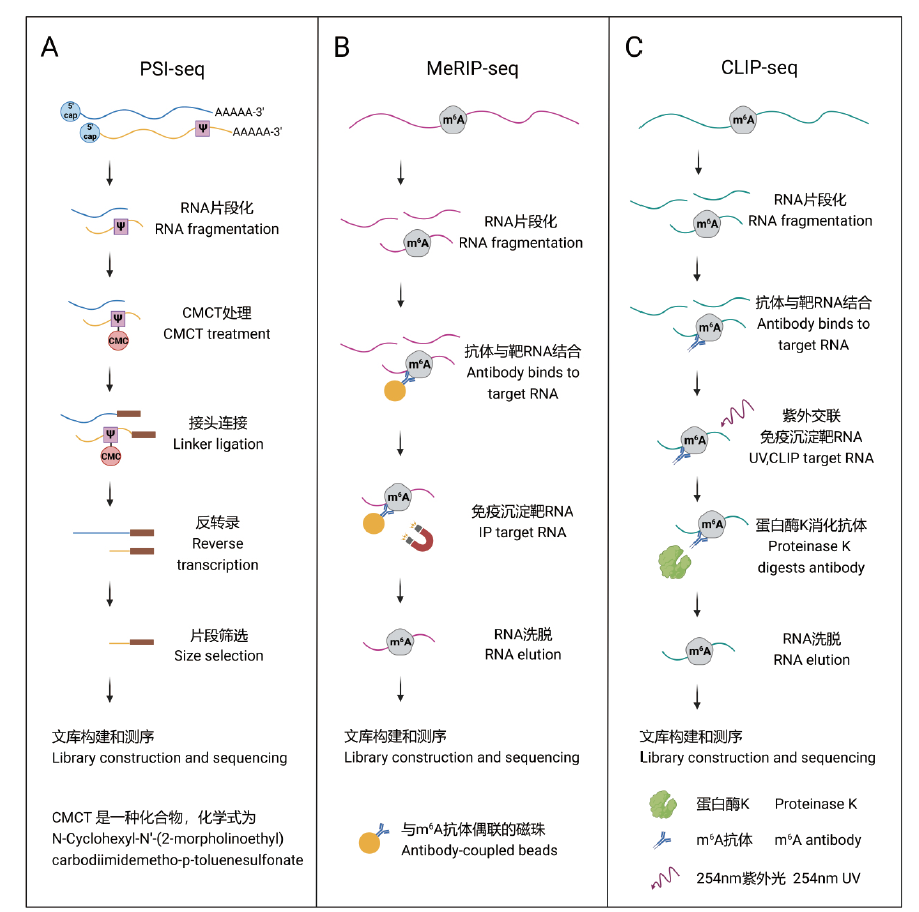
Fig. 1 Processes for detecting RNA modifications based on next-generation sequencing A, B, and C show the workflows of PSI-Seq[61], MeRIP-Seq[18], and CLIP-seq[63], respectively
| RNA修饰 RNA modification | 秀丽隐杆线虫 C. elegans | RNA修饰酶 RNA modifying enzyme | 智人同源物 Homo sapiens homolog | 作用位点 Action site | 生物学功能 Biological function | 参考文献 Reference |
|---|---|---|---|---|---|---|
| m6A | Writers Readers Erasers | METT-10 F33A8.4 C38D4.9 未知 未知 | METTL16 ZCCHC4 METTL5 | snRNA rRNA rRNA | 调控生殖发育 调控寿命 维持应激性 | [ [ [ |
| m5C | Writers Readers Erasers | NSUN-1 NSUN-2 NSUN-4 NSUN-5 RNP-4 SQD-1 PAB-2 未知 | NOP2 NSUN-2 NSUN-4 NSUN-5 RBM8A HNRNPAB PABPC1 | rRNA tRNA rRNA、tRNA rRNA tRNA tRNA tRNA | 调控生长发育 未知 未知 调控寿命 未知 未知 未知 | [ [ [ [ [ [ [ |
| m1A | Writers Readers Erasers | RRAM-1 未知 未知 | NML | rRNA | 调控寿命 | [ |
| Ψ | Writers Readers Erasers | W06H3.2 C50C3.14 未知 未知 | PUS1 SCARNA15 | tRNA snRNA | 未知 未知 | [ [ |
| ac4C | Writers Readers Erasers | NATH-10 未知 未知 | NAT10 | tRNA | 调控生殖发育 | [ |
| Nm | Writers Readers Erasers | HENN-1 未知 未知 | HENMT1 | piRNA | 调控生殖发育 | [ |
| m7G | Writers Readers Erasers | C27F2.4 未知 未知 | BUD23 | rRNA | 调控生长发育 | [ |
Table 1 Overview of common RNA modifications in C. elegans
| RNA修饰 RNA modification | 秀丽隐杆线虫 C. elegans | RNA修饰酶 RNA modifying enzyme | 智人同源物 Homo sapiens homolog | 作用位点 Action site | 生物学功能 Biological function | 参考文献 Reference |
|---|---|---|---|---|---|---|
| m6A | Writers Readers Erasers | METT-10 F33A8.4 C38D4.9 未知 未知 | METTL16 ZCCHC4 METTL5 | snRNA rRNA rRNA | 调控生殖发育 调控寿命 维持应激性 | [ [ [ |
| m5C | Writers Readers Erasers | NSUN-1 NSUN-2 NSUN-4 NSUN-5 RNP-4 SQD-1 PAB-2 未知 | NOP2 NSUN-2 NSUN-4 NSUN-5 RBM8A HNRNPAB PABPC1 | rRNA tRNA rRNA、tRNA rRNA tRNA tRNA tRNA | 调控生长发育 未知 未知 调控寿命 未知 未知 未知 | [ [ [ [ [ [ [ |
| m1A | Writers Readers Erasers | RRAM-1 未知 未知 | NML | rRNA | 调控寿命 | [ |
| Ψ | Writers Readers Erasers | W06H3.2 C50C3.14 未知 未知 | PUS1 SCARNA15 | tRNA snRNA | 未知 未知 | [ [ |
| ac4C | Writers Readers Erasers | NATH-10 未知 未知 | NAT10 | tRNA | 调控生殖发育 | [ |
| Nm | Writers Readers Erasers | HENN-1 未知 未知 | HENMT1 | piRNA | 调控生殖发育 | [ |
| m7G | Writers Readers Erasers | C27F2.4 未知 未知 | BUD23 | rRNA | 调控生长发育 | [ |
| [1] |
He C. Grand challenge commentary: RNA epigenetics?[J]. Nat Chem Biol, 2010, 6(12): 863-865.
doi: 10.1038/nchembio.482 pmid: 21079590 |
| [2] |
Nombela P, Miguel-López B, Blanco S. The role of m6A, m5C and Ψ RNA modifications in cancer: novel therapeutic opportunities[J]. Mol Cancer, 2021, 20: 18.
doi: 10.1186/s12943-020-01263-w pmid: 33461542 |
| [3] |
Schwartz S, Agarwala SD, Mumbach MR, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis[J]. Cell, 2013, 155(6): 1409-1421.
doi: 10.1016/j.cell.2013.10.047 pmid: 24269006 |
| [4] |
Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells[J]. J Mol Biol, 1977, 115(4): 695-714.
doi: 10.1016/0022-2836(77)90110-3 pmid: 592376 |
| [5] |
Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures[J]. J Virol, 1976, 20(1): 45-53.
pmid: 1086370 |
| [6] |
Bruenger E, Kowalak JA, Kuchino Y, et al. 5S rRNA modification in the hyperthermophilic Archaea Sulfolobus solfataricus and Pyrodictium occultum[J]. FASEB J, 1993, 7(1): 196-200.
pmid: 8422966 |
| [7] |
Frye M, Jaffrey SR, Pan T, et al. RNA modifications: what have we learned and where are we headed?[J]. Nat Rev Genet, 2016, 17(6): 365-372.
doi: 10.1038/nrg.2016.47 pmid: 27140282 |
| [8] | Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark[J]. Wiley Interdiscip Rev RNA, 2019, 10(1): e1510. |
| [9] |
Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update[J]. Nucleic Acids Res, 2011, 39(Database issue): D195-D201.
doi: 10.1093/nar/gkq1028 URL |
| [10] |
Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate[J]. Nat Rev Genet, 2017, 18(5): 275-291.
doi: 10.1038/nrg.2016.169 pmid: 28216634 |
| [11] |
Yang Y, Sun BF, Xiao W, et al. Dynamic m6A modification and its emerging regulatory role in mRNA splicing[J]. Sci Bull, 2015, 60(1): 21-32.
doi: 10.1007/s11434-014-0695-6 URL |
| [12] | Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation[J]. Nat Rev Mol Cell Biol, 2019, 20(10): 608-624. |
| [13] |
Frye M, Harada BT, Behm M, et al. RNA modifications modulate gene expression during development[J]. Science, 2018, 361(6409): 1346-1349.
doi: 10.1126/science.aau1646 pmid: 30262497 |
| [14] |
Corsi AK, Wightman B, Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans[J]. Genetics, 2015, 200(2): 387-407.
doi: 10.1534/genetics.115.176099 pmid: 26088431 |
| [15] |
Billi AC, Alessi AF, Khivansara V, et al. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs[J]. PLoS Genet, 2012, 8(4): e1002617.
doi: 10.1371/journal.pgen.1002617 URL |
| [16] |
Rong BW, Zhang Q, Wan JK, et al. Ribosome 18S m6A methyltransferase METTL5 promotes translation initiation and breast cancer cell growth[J]. Cell Rep, 2020, 33(12): 108544.
doi: 10.1016/j.celrep.2020.108544 URL |
| [17] |
Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation[J]. Nat Rev Genet, 2014, 15(5): 293-306.
doi: 10.1038/nrg3724 |
| [18] |
Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons[J]. Cell, 2012, 149(7): 1635-1646.
doi: 10.1016/j.cell.2012.05.003 pmid: 22608085 |
| [19] |
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq[J]. Nature, 2012, 485(7397): 201-206.
doi: 10.1038/nature11112 |
| [20] |
Shi HL, Wei JB, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers[J]. Mol Cell, 2019, 74(4): 640-650.
doi: S1097-2765(19)30317-X pmid: 31100245 |
| [21] |
Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism[J]. Cell Res, 2018, 28(6): 616-624.
doi: 10.1038/s41422-018-0040-8 |
| [22] |
Lewis CJT, Pan T, Kalsotra A. RNA modifications and structures cooperate to guide RNA-protein interactions[J]. Nat Rev Mol Cell Biol, 2017, 18(3): 202-210.
doi: 10.1038/nrm.2016.163 |
| [23] |
Li LP, Zang LQ, Zhang FR, et al. Fat mass and obesity-associated(FTO)protein regulates adult neurogenesis[J]. Hum Mol Genet, 2017, 26(13): 2398-2411.
doi: 10.1093/hmg/ddx128 URL |
| [24] |
Yoon KJ, Ringeling FR, Vissers C, et al. Temporal control of mammalian cortical neurogenesis by m6A methylation[J]. Cell, 2017, 171(4): 877-889.e17.
doi: 10.1016/j.cell.2017.09.003 URL |
| [25] |
Xie W, Ma LL, Xu YQ, et al. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism[J]. Biochem Biophys Res Commun, 2019, 518(1): 120-126.
doi: 10.1016/j.bbrc.2019.08.018 URL |
| [26] |
Liu J, Luo GZ, Sun J, et al. METTL14 is essential for β-cell survival and insulin secretion[J]. Biochim Biophys Acta Mol Basis Dis, 2019, 1865(9): 2138-2148.
doi: 10.1016/j.bbadis.2019.04.011 URL |
| [27] |
Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA[J]. Nucleic Acids Res, 2012, 40(11): 5023-5033.
doi: 10.1093/nar/gks144 pmid: 22344696 |
| [28] |
Amort T, Rieder D, Wille A, et al. Distinct 5-methylcytosine profiles in poly(A)RNA from mouse embryonic stem cells and brain[J]. Genome Biol, 2017, 18(1): 1.
doi: 10.1186/s13059-016-1139-1 URL |
| [29] |
Hong B, Brockenbrough JS, Wu P, et al. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast[J]. Mol Cell Biol, 1997, 17(1): 378-388.
doi: 10.1128/MCB.17.1.378 pmid: 8972218 |
| [30] |
Metodiev MD, Spåhr H, Loguercio Polosa P, et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly[J]. PLoS Genet, 2014, 10(2): e1004110.
doi: 10.1371/journal.pgen.1004110 URL |
| [31] |
Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2[J]. Science, 2006, 311(5759): 395-398.
doi: 10.1126/science.1120976 pmid: 16424344 |
| [32] |
Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader[J]. Cell Res, 2017, 27(5): 606-625.
doi: 10.1038/cr.2017.55 pmid: 28418038 |
| [33] |
Fu LJ, Guerrero CR, Zhong N, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA[J]. J Am Chem Soc, 2014, 136(33): 11582-11585.
doi: 10.1021/ja505305z pmid: 25073028 |
| [34] |
Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders[J]. EMBO J, 2014, 33(18): 2020-2039.
doi: 10.15252/embj.201489282 pmid: 25063673 |
| [35] |
Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility[J]. Stem Cell Reports, 2017, 8(1): 112-124.
doi: S2213-6711(16)30276-4 pmid: 28041877 |
| [36] |
Xue SL, Xu H, Sun Z, et al. Depletion of TRDMT1 affects 5-methylcytosine modification of mRNA and inhibits HEK293 cell proliferation and migration[J]. Biochem Biophys Res Commun, 2019, 520(1): 60-66.
doi: 10.1016/j.bbrc.2019.09.098 URL |
| [37] |
Sun Z, Xue SL, Xu H, et al. Effects of NSUN2 deficiency on the mRNA 5-methylcytosine modification and gene expression profile in HEK293 cells[J]. Epigenomics, 2019, 11(4): 439-453.
doi: 10.2217/epi-2018-0169 pmid: 30526041 |
| [38] |
Li XY, Xiong XS, Wang K, et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome[J]. Nat Chem Biol, 2016, 12(5): 311-316.
doi: 10.1038/nchembio.2040 |
| [39] |
Safra M, Sas-Chen A, Nir R, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution[J]. Nature, 2017, 551(7679): 251-255.
doi: 10.1038/nature24456 URL |
| [40] |
Ueda Y, Ooshio I, Fusamae Y, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells[J]. Sci Rep, 2017, 7: 42271.
doi: 10.1038/srep42271 pmid: 28205560 |
| [41] |
Liu FG, Clark W, Luo GZ, et al. ALKBH1-mediated tRNA demethylation regulates translation[J]. Cell, 2016, 167(3): 816-828.e16.
doi: S0092-8674(16)31323-X pmid: 27745969 |
| [42] |
Dai XX, Wang TL, Gonzalez G, et al. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA[J]. Anal Chem, 2018, 90(11): 6380-6384.
doi: 10.1021/acs.analchem.8b01703 URL |
| [43] |
Wang Y, Huang Q, Deng T, et al. Clinical significance of TRMT6 in hepatocellular carcinoma: a bioinformatics-based study[J]. Med Sci Monit, 2019, 25: 3894-3901.
doi: 10.12659/MSM.913556 URL |
| [44] |
Hartl M. The quest for targets executing MYC-dependent cell transformation[J]. Front Oncol, 2016, 6: 132.
doi: 10.3389/fonc.2016.00132 pmid: 27313991 |
| [45] |
Zhao YS, Zhao QJ, Kaboli PJ, et al. m1A regulated genes modulate PI3K/AKT/mTOR and ErbB pathways in gastrointestinal cancer[J]. Transl Oncol, 2019, 12(10): 1323-1333.
doi: S1936-5233(19)30192-5 pmid: 31352195 |
| [46] |
Xue C, Chu QF, Zheng QX, et al. Role of main RNA modifications in cancer: N6-methyladenosine, 5-methylcytosine, and pseudouridine[J]. Sig Transduct Target Ther, 2022, 7: 142.
doi: 10.1038/s41392-022-01003-0 |
| [47] |
Karikó K, Muramatsu H, Welsh FA, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability[J]. Mol Ther, 2008, 16(11): 1833-1840.
doi: 10.1038/mt.2008.200 pmid: 18797453 |
| [48] |
Zeleznik OA, Eliassen AH, Kraft P, et al. A prospective analysis of circulating plasma metabolites associated with ovarian cancer risk[J]. Cancer Res, 2020, 80(6): 1357-1367.
doi: 10.1158/0008-5472.CAN-19-2567 pmid: 31969373 |
| [49] |
Jin GH, Xu MQ, Zou MS, et al. The processing, gene regulation, biological functions, and clinical relevance of N4-acetylcytidine on RNA: a systematic review[J]. Mol Ther Nucleic Acids, 2020, 20: 13-24.
doi: 10.1016/j.omtn.2020.01.037 URL |
| [50] |
Karthiya R, Wasil SM, Khandelia P. Emerging role of N4-acetylcytidine modification of RNA in gene regulation and cellular functions[J]. Mol Biol Rep, 2020, 47(11): 9189-9199.
doi: 10.1007/s11033-020-05963-w pmid: 33174082 |
| [51] |
Arango D, Sturgill D, Alhusaini N, et al. Acetylation of cytidine in mRNA promotes translation efficiency[J]. Cell, 2018, 175(7): 1872-1886.e24.
doi: S0092-8674(18)31383-7 pmid: 30449621 |
| [52] |
Orita I, Futatsuishi R, Adachi K, et al. Random mutagenesis of a hyperthermophilic archaeon identified tRNA modifications associated with cellular hyperthermotolerance[J]. Nucleic Acids Res, 2019, 47(4): 1964-1976.
doi: 10.1093/nar/gky1313 pmid: 30605516 |
| [53] |
Tsai K, Jaguva Vasudevan AA, Martinez Campos C, et al. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability[J]. Cell Host Microbe, 2020, 28(2): 306-312.e6.
doi: S1931-3128(20)30293-6 pmid: 32533923 |
| [54] |
Motorin Y, Marchand V. Analysis of RNA modifications by second- and third-generation deep sequencing: 2020 update[J]. Genes, 2021, 12(2): 278.
doi: 10.3390/genes12020278 URL |
| [55] |
Nees G, Kaufmann A, Bauer S. Detection of RNA modifications by HPLC analysis and competitive ELISA[J]. Methods Mol Biol, 2014, 1169: 3-14.
doi: 10.1007/978-1-4939-0882-0_1 pmid: 24957224 |
| [56] | Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension[J]. Methods Enzymol, 1988, 164: 481-489. |
| [57] |
Zhao LY, Song JH, Liu YB, et al. Mapping the epigenetic modifications of DNA and RNA[J]. Protein Cell, 2020, 11(11): 792-808.
doi: 10.1007/s13238-020-00733-7 |
| [58] |
Motorin Y, Helm M. Methods for RNA modification mapping using deep sequencing: established and new emerging technologies[J]. Genes, 2019, 10(1): 35.
doi: 10.3390/genes10010035 URL |
| [59] |
Linder B, Jaffrey SR. Discovering and mapping the modified nucleotides that comprise the epitranscriptome of mRNA[J]. Cold Spring Harb Perspect Biol, 2019, 11(6): a032201.
doi: 10.1101/cshperspect.a032201 URL |
| [60] |
Krogh N, Nielsen H. Sequencing-based methods for detection and quantitation of ribose methylations in RNA[J]. Methods, 2019, 156: 5-15.
doi: S1046-2023(18)30171-3 pmid: 30503826 |
| [61] |
Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae[J]. PLoS One, 2014, 9(10): e110799.
doi: 10.1371/journal.pone.0110799 URL |
| [62] |
Yang L, Chen X, Qian X, et al. Comprehensive analysis of the transcriptome-wide m6A methylome in endometrioid ovarian cancer[J]. Front Oncol, 2022, 12: 844613.
doi: 10.3389/fonc.2022.844613 URL |
| [63] |
Ke SD, Alemu EA, Mertens C, et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation[J]. Genes Dev, 2015, 29(19): 2037-2053.
doi: 10.1101/gad.269415.115 URL |
| [64] |
Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years[J]. Nat Rev Genet, 2019, 20(11): 631-656.
doi: 10.1038/s41576-019-0150-2 pmid: 31341269 |
| [65] |
Garalde DR, Snell EA, Jachimowicz D, et al. Highly parallel direct RNA sequencing on an array of nanopores[J]. Nat Methods, 2018, 15(3): 201-206.
doi: 10.1038/nmeth.4577 pmid: 29334379 |
| [66] |
Rhoads A, Au KF. PacBio sequencing and its applications[J]. Genomics Proteomics Bioinformatics, 2015, 13(5): 278-289.
doi: 10.1016/j.gpb.2015.08.002 URL |
| [67] |
Li RS, Ren XL, Ding QT, et al. Direct full-length RNA sequencing reveals unexpected transcriptome complexity during Caenorhabditis elegans development[J]. Genome Res, 2020, 30(2): 287-298.
doi: 10.1101/gr.251512.119 URL |
| [68] |
Vilfan ID, Tsai YC, Clark TA, et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription[J]. J Nanobiotechnology, 2013, 11: 8.
doi: 10.1186/1477-3155-11-8 |
| [69] |
Sendinc E, Valle-Garcia D, Jiao AL, et al. Analysis of m6A RNA methylation in Caenorhabditis elegans[J]. Cell Discov, 2020, 6: 47.
doi: 10.1038/s41421-020-00186-6 pmid: 32695436 |
| [70] |
Pendleton KE, Chen BB, Liu KQ, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention[J]. Cell, 2017, 169(5): 824-835.e14.
doi: S0092-8674(17)30530-5 pmid: 28525753 |
| [71] |
Dorsett M, Westlund B, Schedl T. METT-10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans[J]. Genetics, 2009, 183(1): 233-247.
doi: 10.1534/genetics.109.105270 pmid: 19596901 |
| [72] |
Mendel M, Delaney K, Pandey RR, et al. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing[J]. Cell, 2021, 184(12): 3125-3142.e25.
doi: 10.1016/j.cell.2021.03.062 URL |
| [73] |
Hansen M, Taubert S, Crawford D, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans[J]. Aging Cell, 2007, 6(1): 95-110.
doi: 10.1111/ace.2007.6.issue-1 URL |
| [74] |
Liberman N, O'Brown ZK, Earl AS, et al. N6-adenosine methylation of ribosomal RNA affects lipid oxidation and stress resistance[J]. Sci Adv, 2020, 6(17): eaaz4370.
doi: 10.1126/sciadv.aaz4370 URL |
| [75] |
Adamla F, Rollins J, Newsom M, et al. A novel Caenorhabditis elegans proteinopathy model shows changes in mRNA translational frameshifting during aging[J]. Cell Physiol Biochem, 2019, 52(5): 970-983.
doi: 10.33594/000000000 URL |
| [76] |
Heissenberger C, Rollins JA, Krammer TL, et al. The ribosomal RNA m5C methyltransferase NSUN-1 modulates healthspan and oogenesis in Caenorhabditis elegans[J]. eLife, 2020, 9: e56205.
doi: 10.7554/eLife.56205 URL |
| [77] |
Rollins JA, Howard AC, Dobbins SK, et al. Assessing health span in Caenorhabditis elegans: lessons from short-lived mutants[J]. J Gerontol A Biol Sci Med Sci, 2017, 72(4): 473-480.
doi: 10.1093/gerona/glw248 URL |
| [78] | Bansal A, Zhu LJ, Yen K, et al. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants[J]. Proc Natl Acad Sci USA, 2015, 112(3): E277-E286. |
| [79] |
Navarro IC, Tuorto F, Jordan D, et al. Translational adaptation to heat stress is mediated by RNA 5-methylcytosine in Caenorhabditis elegans[J]. EMBO J, 2021, 40(6): e105496.
doi: 10.15252/embj.2020105496 URL |
| [80] |
Navarro IC, Suen KM, Bensaddek D, et al. Identification of putative reader proteins of 5-methylcytosine and its derivatives in Caenorhabditis elegans RNA[J]. bioRxiv, 2022, DOI:10.1101/2022.01.24.477592.
doi: 10.1101/2022.01.24.477592 |
| [81] | Shima H, Igarashi K. N1-methyladenosine(m1A)RNA modification: the key to ribosome control[J]. J Biochem, 2020, 167(6): 535-539. |
| [82] |
Yokoyama W, Hirota K, Wan HH, et al. rRNA adenine methylation requires T07A9.8 gene as rram-1 in Caenorhabditis elegans[J]. J Biochem, 2018, 163(6): 465-474.
doi: 10.1093/jb/mvy018 pmid: 29385568 |
| [83] |
Patton JR, Padgett RW. Caenorhabditis elegans pseudouridine synthase 1 activity in vivo: tRNA is a substrate, but not U2 small nuclear RNA[J]. Biochem J, 2003, 372(Pt 2): 595-602.
doi: 10.1042/bj20021938 URL |
| [84] |
Deryusheva S, Gall JG. Orchestrated positioning of post-transcriptional modifications at the branch point recognition region of U2 snRNA[J]. RNA, 2018, 24(1): 30-42.
doi: 10.1261/rna.063842.117 pmid: 28974555 |
| [85] |
Patton JR, Padgett RW. Pseudouridine modification in Caenorhabditis elegans spliceosomal snRNAs: unique modifications are found in regions involved in snRNA-snRNA interactions[J]. BMC Mol Biol, 2005, 6: 20.
doi: 10.1186/1471-2199-6-20 |
| [86] |
Bortolin-Cavaillé ML, Quillien A, Thalalla Gamage S, et al. Probing small ribosomal subunit RNA helix 45 acetylation across eukaryotic evolution[J]. Nucleic Acids Res, 2022, 50(11): 6284-6299.
doi: 10.1093/nar/gkac404 pmid: 35648437 |
| [87] |
Hintze M, Katsanos D, Shahrezaei V, et al. Phenotypic robustness of epidermal stem cell number in C. elegans is modulated by the activity of the conserved N-acetyltransferase nath-10/NAT10[J]. Front Cell Dev Biol, 2021, 9: 640856.
doi: 10.3389/fcell.2021.640856 URL |
| [88] |
Duveau F, Félix MA. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans[J]. PLoS Biol, 2012, 10(1): e1001230.
doi: 10.1371/journal.pbio.1001230 URL |
| [89] |
Svendsen JM, Reed KJ, Vijayasarathy T, et al. Henn-1/HEN1 promotes germline immortality in Caenorhabditis elegans[J]. Cell Rep, 2019, 29(10): 3187-3199.e4.
doi: S2211-1247(19)31447-0 pmid: 31801082 |
| [90] |
Pastore B, Hertz HL, Price IF, et al. Pre-PiRNA trimming and 2'-O-methylation protect piRNAs from 3' tailing and degradation in C. elegans[J]. Cell Rep, 2021, 36(9): 109640.
doi: 10.1016/j.celrep.2021.109640 URL |
| [91] |
Zhu CM, Yan Q, Weng CC, et al. Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA[J]. Proc Natl Acad Sci USA, 2018, 115(40): 10082-10087.
doi: 10.1073/pnas.1800974115 pmid: 30224484 |
| [1] | ZHAO Jin-ling, AN Lei, REN Xiao-liang. Development of Single Cell Transcriptome Sequencing Technology and Its Application in Caenorhabditis elegans [J]. Biotechnology Bulletin, 2023, 39(6): 158-170. |
| [2] | LONG Wen-lin, GUO Hui, SHENG Jie, SONG Ru-hui, XU Yao. Role of m6A RNA Methylation in Tumorigenesis and Development [J]. Biotechnology Bulletin, 2019, 35(6): 178-186. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||