








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 39-46.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0334
Previous Articles Next Articles
Received:2024-04-09
Online:2024-08-26
Published:2024-09-05
Contact:
QI Ji
E-mail:r_wang21@m.fudan.edu.cn;qij@fudan.edu.cn
WANG Rui, QI Ji. Integrating Histological Image Information to Enhance Cell Clustering Resolution in Spatial Transcriptome[J]. Biotechnology Bulletin, 2024, 40(8): 39-46.
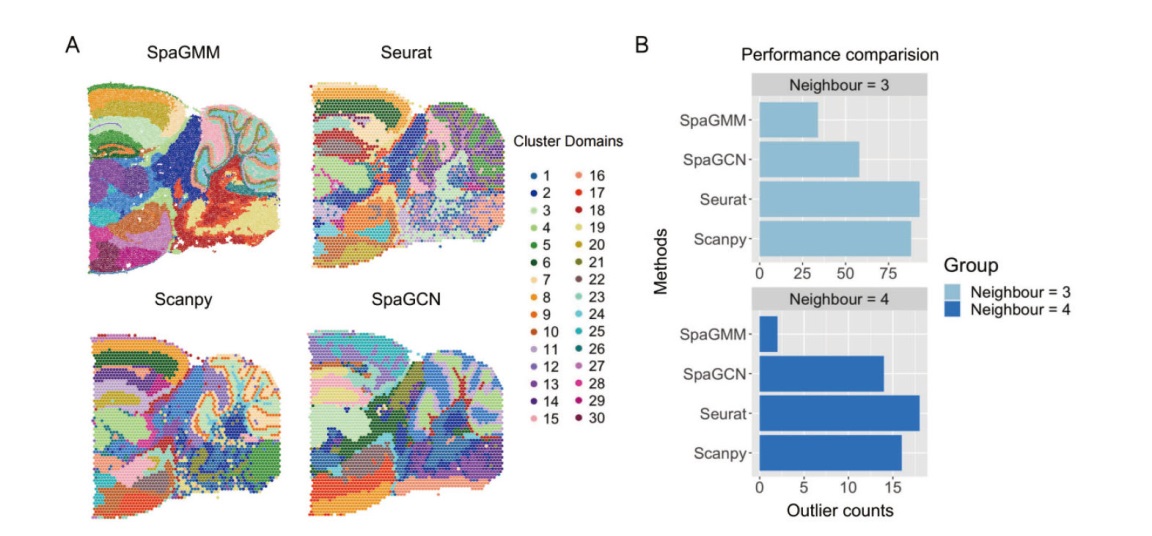
Fig. 1 Performance comparison of the dataset of posterior section of mouse brain on 10X Visium platform by four methods A: Illustration of clustering results(30 groups)of posterior section of mouse brain by each method. B: Outlier numbers in clustering results by the each method
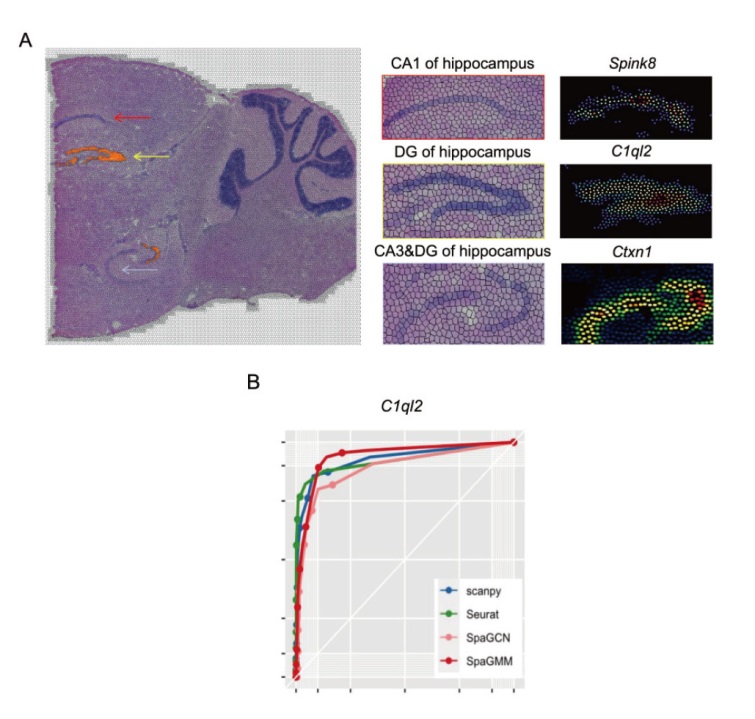
Fig. 2 Reconstruction of gene spatial expression pattern of SpaGMM in the hippocampus region of mouse brain and classification result A: Comparison of spatial expressions of marker genes in the CA1,CA3 and DG regions of hippocampus. B: Evaluation of the four algorithms on the classification of dentate gyrus type neurons according to spatial expression of marker gene C1ql2
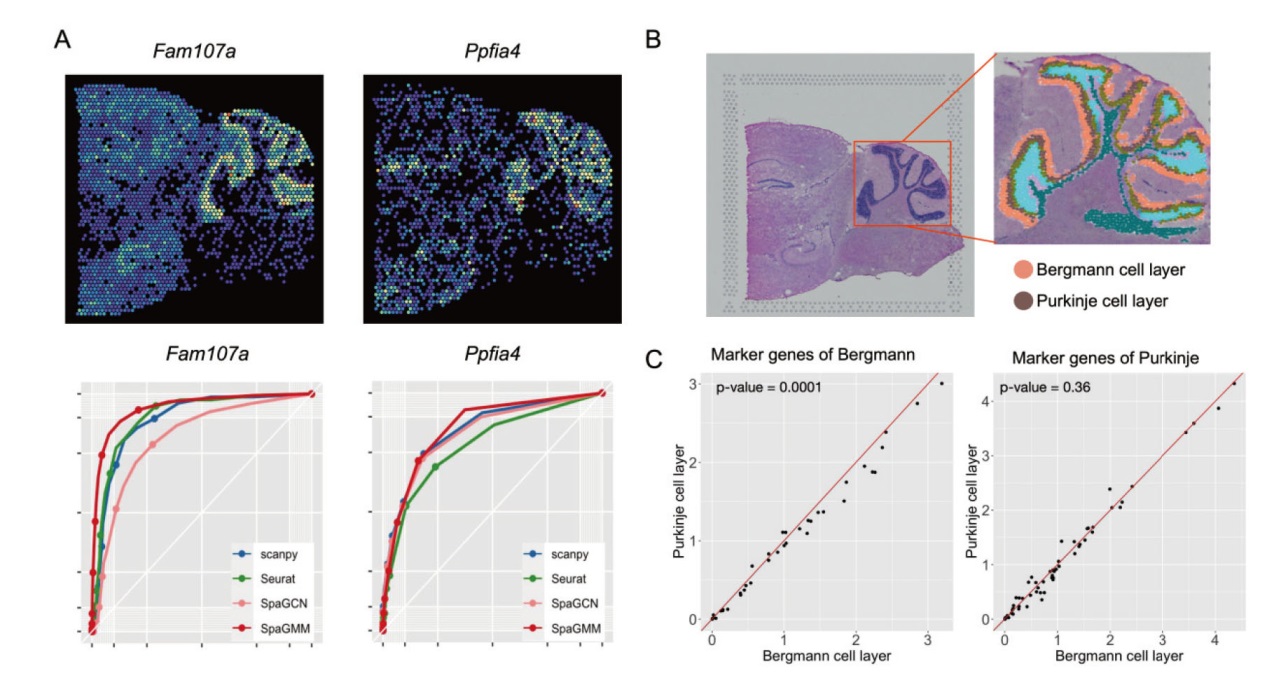
Fig. 3 Comparison of classification results on the cerebellum of mouse brain by four methods A: Spatial expression pattern of marker genes at molecular layer and particle layer, and evaluation of the classification results of the four methods. B: Illustration of different layers on cerebellum recognized by SpaGMM. C: Illustration of expression of marker genes for Purkinje and Bergman cells in different spatial domains
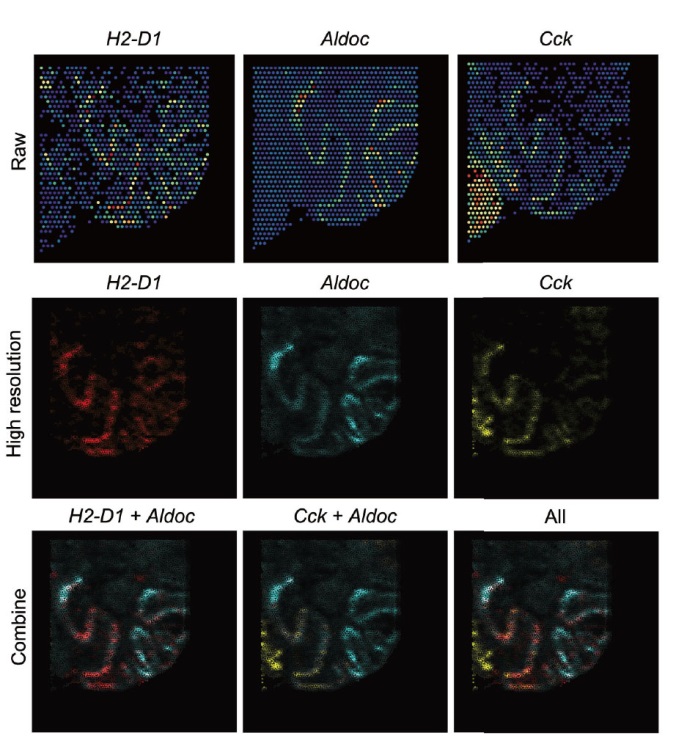
Fig. 4 Spatial expression patterns of Aldoc(Zebrin II positive)and Cck, H2-D1(Zebrin II negative)genes in Purkinje cell layer From top to bottom, the images indicate the heat map of original gene expression(the color from blue to red indicates the gene expressions from low to high), the heat map of SpaGMM reconstructed super-resolution gene expression(the color from light to deep indicates the gene expressions from low to high), and the integration of multiple genes shows the complementary expression pattern in the Purkinje cell layer
| [1] | Lee J, Hyeon DY, Hwang D. Single-cell multiomics: technologies and data analysis methods[J]. Exp Mol Med, 2020, 52(9): 1428-1442. |
| [2] | Ahmed R, Zaman T, Chowdhury F, et al. Single-cell RNA sequencing with spatial transcriptomics of cancer tissues[J]. Int J Mol Sci, 2022, 23(6): 3042. |
| [3] | Lei YL, Tang R, Xu J, et al. Applications of single-cell sequencing in cancer research: progress and perspectives[J]. J Hematol Oncol, 2021, 14(1): 91. |
| [4] |
Moses L, Pachter L. Museum of spatial transcriptomics[J]. Nat Methods, 2022, 19(5): 534-546.
doi: 10.1038/s41592-022-01409-2 pmid: 35273392 |
| [5] | Lake BB, Menon R, Winfree S, et al. An atlas of healthy and injured cell states and niches in the human kidney[J]. Nature, 2023, 619(7970): 585-594. |
| [6] | Galeano Niño JL, Wu HR, LaCourse KD, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer[J]. Nature, 2022, 611(7937): 810-817. |
| [7] | Li QY, Zhang XY, Ke RQ. Spatial transcriptomics for tumor heterogeneity analysis[J]. Front Genet, 2022, 13: 906158. |
| [8] | Mazzei M, Vascellari M, Zanardello C, et al. Quantitative real time polymerase chain reaction(RT-qPCR)and RNAscope in situ hybridization(RNA-ISH)as effective tools to diagnose feline herpesvirus-1-associated dermatitis[J]. Vet Dermatol, 2019, 30(6): 491. |
| [9] | Eng CH L, Lawson M, Zhu Q, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH[J]. Nature, 2019, 568(7751): 235-239. |
| [10] | Zhou XY, Karpur A, Luo LJ, et al. StarMap for category-agnostic keypoint and viewpoint estimation[C]// European Conference on Computer Vision. Cham: Springer, 2018: 328-345. |
| [11] | Fürth D, Hatini V, Lee JH. In situ transcriptome accessibility sequencing(INSTA-seq)[J]. bioRxiv, 2019. DOI: 10.1101/722819. |
| [12] |
Stickels RR, Murray E, Kumar P, et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2[J]. Nat Biotechnol, 2021, 39(3): 313-319.
doi: 10.1038/s41587-020-0739-1 pmid: 33288904 |
| [13] | Kephart M. A 10x visium approach: a spatial RNA-Seq analysis of renal tissue in Peromyscus eremicus[D]. ProQuest Dissertations Publishing, 2023. |
| [14] | Luoma J, Nastou K, Ohta T, et al. S1000: a better taxonomic Name corpus for biomedical information extraction[J]. Bioinformatics, 2023, 39(6): btad369. |
| [15] | Xia KK, Sun HX, Li J, et al. The single-cell stereo-seq reveals region-specific cell subtypes and transcriptome profiling in Arabidopsis leaves[J]. Dev Cell, 2022, 57(10): 1299-1310.e4. |
| [16] | Su G, Qin XY, Enninful A, et al. Spatial multi-omics sequencing for fixed tissue via DBiT-seq[J]. STAR Protoc, 2021, 2(2): 100532. |
| [17] |
Satija R, Farrell JA, Gennert D, et al. Spatial reconstruction of single-cell gene expression data[J]. Nat Biotechnol, 2015, 33(5): 495-502.
doi: 10.1038/nbt.3192 pmid: 25867923 |
| [18] |
Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis[J]. Genome Biol, 2018, 19(1): 15.
doi: 10.1186/s13059-017-1382-0 pmid: 29409532 |
| [19] |
Hu J, Li XJ, Coleman K, et al. SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network[J]. Nat Methods, 2021, 18(11): 1342-1351.
doi: 10.1038/s41592-021-01255-8 pmid: 34711970 |
| [20] |
Zhao E, Stone MR, Ren X, et al. Spatial transcriptomics at subspot resolution with BayesSpace[J]. Nat Biotechnol, 2021, 39(11): 1375-1384.
doi: 10.1038/s41587-021-00935-2 pmid: 34083791 |
| [21] |
Scrucca L, Fop M, Murphy TB, et al. Mclust 5: clustering, classification and density estimation using Gaussian finite mixture models[J]. R J, 2016, 8(1): 289-317.
pmid: 27818791 |
| [22] |
Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities[J]. Sci Rep, 2019, 9(1): 5233.
doi: 10.1038/s41598-019-41695-z pmid: 30914743 |
| [23] | Ahmed M, Seraj R, Islam SMS. The k-means algorithm: a comprehensive survey and performance evaluation[J]. Electronics, 2020, 9(8): 1295. |
| [24] | Pham D, Tan X, Xu J, et al. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues[J]. bioRxiv, 2020. DOI: 10.1101/2020.05.31.125658. |
| [25] | Pan V, Schreiber R. An improved Newton iteration for the generalized inverse of a matrix, with applications[J]. SIAM J Sci Stat Comput, 1991, 12(5): 1109-1130. |
| [26] | Chung MK. Gaussian kernel smoothing[EB/OL]. 2020: arXiv: 2007.09539. http://arxiv.org/abs/2007.09539. |
| [27] | Achanta R, Shaji A, Smith K, et al. SLIC superpixels compared to state-of-the-art superpixel methods[J]. IEEE Trans Pattern Anal Mach Intell, 2012, 34(11): 2274-2282. |
| [28] | Hurtik P, Madrid N. Bilinear interpolation over fuzzified images: enlargement[C]// 2015 IEEE International Conference on Fuzzy Systems(FUZZ-IEEE). Istanbul, Turkey. Piscataway, NJ: IEEE, 2015: 1-8. |
| [29] | Reynolds D. Gaussian mixture models[M]// Li SZ, Jain A. Encyclopedia of Biometrics. Bostonm MA: Springer, 2009: 659-663. |
| [30] | De Meo P, Ferrara E, Fiumara G, et al. Generalized louvain method for community detection in large networks[C]// 2011 11th International Conference on Intelligent Systems Design and Applications, 2011: 88-93. |
| [31] |
Xu C, Jin XY, Wei SR, et al. DeepST: identifying spatial domains in spatial transcriptomics by deep learning[J]. Nucleic Acids Res, 2022, 50(22): e131.
doi: 10.1093/nar/gkac901 pmid: 36250636 |
| [32] | Chen YM, Zhou SY, Li M, et al. STEEL enables high-resolution delineation of spatiotemporal transcriptomic data[J]. Brief Bioinform, 2023, 24(2): bbad068. |
| [33] | Franzén O, Gan LM, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data[J]. Database, 2019, 2019: baz046. |
| [34] | Perkel DJ, Hestrin S, Sah P, et al. Excitatory synaptic currents in Purkinje cells[J]. Proc R Soc Lond B, 1990, 241(1301): 116-121. |
| [35] | Lin YC, Hsu CC H, Wang PN, et al. The relationship between zebrin expression and cerebellar functions: insights from neuroimaging studies[J]. Front Neurol, 2020, 11: 315. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
