








生物技术通报 ›› 2021, Vol. 37 ›› Issue (10): 169-178.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1474
收稿日期:2020-12-02
出版日期:2021-10-26
发布日期:2021-11-12
作者简介:高竞溪,女,研究方向:抗体免疫;E-mail: 基金资助:
GAO Jing-xi( ), GAO Ke-xing, LU Fei, JI Feng, GUO Zhi-gang(
), GAO Ke-xing, LU Fei, JI Feng, GUO Zhi-gang( )
)
Received:2020-12-02
Published:2021-10-26
Online:2021-11-12
摘要:
综合预测可与SARS-CoV发生交叉免疫反应的SARS-CoV-2 棘突蛋白(S蛋白)优势B细胞抗原表位,为开发SARS-CoV-2和SARS-CoV S蛋白通用表位疫苗和相关单抗奠定基础。采用MEGA5.05中的Neighbor-joining法构建基于SARS-CoV-2 S蛋白氨基酸序列的系统发生树;参考SARS-CoV-2毒株与SARS-CoV毒株S蛋白氨基酸序列比对及跨膜分析结果,筛选出二者可能存在的共同抗原表位区段(944-1 213aa)。以SARS-CoV-2 Wuhan-Hu-1株为研究对象,其目标区段的亲水性指数、柔性区段、蛋白质表面可能性和抗原指数等参数由DNAStar软件中的Protean模块进行分析预测;结合Phyre2在线工具模拟的空间结构,综合预测可与SARS-CoV发生交叉免疫反应的SARS-CoV-2 S蛋白B细胞优势抗原表位。SARS-CoV-2 S蛋白氨基酸序列与SARS-CoV S蛋白具有最高相似性(77.5%),与其它可感染人类的冠状病毒(MERS-CoV、HCoV-HKU1、HCoV-NL63、HCoV-229E)S蛋白存在显著差异,相似性均低于30.3%。Wuhan-Hu-1株S蛋白具有3个疏水核心,可能存在较强的可变性。目标B细胞抗原表位大概率分布于959-966、973-979、1 003-1 011、1 030-1 037、1 057-1 070、1 079-1 085、1 123-1 132、1 174-1 179位氨基酸区段。预测的SARS-CoV-2和SARS-CoV S蛋白共同抗原表位可为通用表位疫苗的设计、单抗的制备、相关药物的快速筛选等工作提供参考。
高竞溪, 高珂星, 鲁非, 纪锋, 郭志刚. 与SARS-CoV有交叉免疫反应的SARS-CoV-2 S蛋白B细胞抗原表位预测[J]. 生物技术通报, 2021, 37(10): 169-178.
GAO Jing-xi, GAO Ke-xing, LU Fei, JI Feng, GUO Zhi-gang. Prediction of SARS-CoV-2 S Protein B Cell Antigenic Epitope Cross-immunizing with SARS-CoV[J]. Biotechnology Bulletin, 2021, 37(10): 169-178.
| 毒株Isolate | 序列号Accession no. | 宿主Host | 国家或地区Country or area |
|---|---|---|---|
| WHU01 | QHD43416.1 | Human | China |
| WHU02 | QHO62112.1 | Human | China |
| Peru-10 | QIS60288.1 | Human | Peru |
| SP02 | QIG55994.1 | Human | Brazil |
| NC0025 | QJA17752.1 | Human | USA:NC |
| MA3653 | QIZ16497.1 | Human | USA:MA |
| CHUKtc55NS | ABD73002.1 | Bat | China:Hong Kong |
| CHUKtc51L | ABD73000.1 | Bat | China:Hong Kong |
| CHUKtc46NP | ABD72997.1 | Bat | China:Hong Kong |
| ExoN1 | AGT21273.1 | Human | USA:Nashville,TN |
| TC3 | AAP97882.1 | Human | China:Taiwan |
| QXC1 | AAR86788.1 | Human | China |
| NV2020 | AVN89453.1 | Camelus dromedarius | Nigeria |
| CIRAD-HKU785 | AVN89344.1 | Camelus dromedarius | Burkina Faso |
| CIRAD-HKU213 | AVN89324.1 | Camelus dromedarius | Morocco |
| AAU-EPHI-HKU4458 | AVN89313.1 | Camelus dromedarius | Ethiopia |
| BN1 | AOG74783.1 | Human | Germany |
| SC9724 | QEO75985.1 | Human | USA |
| SC0768 | QEG03748.1 | Human | USA |
| SI17244 | AYN64561.1 | Human | Thailand |
| BJ01-p9 | AMN88694.1 | Human | China |
| BJ01-p3 | AMN88686.1 | Human | China |
| Caen1 | ADN03339.1 | Human | France |
表1 研究所用的冠状病毒株
Table 1 Coronavirus strains used in this study
| 毒株Isolate | 序列号Accession no. | 宿主Host | 国家或地区Country or area |
|---|---|---|---|
| WHU01 | QHD43416.1 | Human | China |
| WHU02 | QHO62112.1 | Human | China |
| Peru-10 | QIS60288.1 | Human | Peru |
| SP02 | QIG55994.1 | Human | Brazil |
| NC0025 | QJA17752.1 | Human | USA:NC |
| MA3653 | QIZ16497.1 | Human | USA:MA |
| CHUKtc55NS | ABD73002.1 | Bat | China:Hong Kong |
| CHUKtc51L | ABD73000.1 | Bat | China:Hong Kong |
| CHUKtc46NP | ABD72997.1 | Bat | China:Hong Kong |
| ExoN1 | AGT21273.1 | Human | USA:Nashville,TN |
| TC3 | AAP97882.1 | Human | China:Taiwan |
| QXC1 | AAR86788.1 | Human | China |
| NV2020 | AVN89453.1 | Camelus dromedarius | Nigeria |
| CIRAD-HKU785 | AVN89344.1 | Camelus dromedarius | Burkina Faso |
| CIRAD-HKU213 | AVN89324.1 | Camelus dromedarius | Morocco |
| AAU-EPHI-HKU4458 | AVN89313.1 | Camelus dromedarius | Ethiopia |
| BN1 | AOG74783.1 | Human | Germany |
| SC9724 | QEO75985.1 | Human | USA |
| SC0768 | QEG03748.1 | Human | USA |
| SI17244 | AYN64561.1 | Human | Thailand |
| BJ01-p9 | AMN88694.1 | Human | China |
| BJ01-p3 | AMN88686.1 | Human | China |
| Caen1 | ADN03339.1 | Human | France |
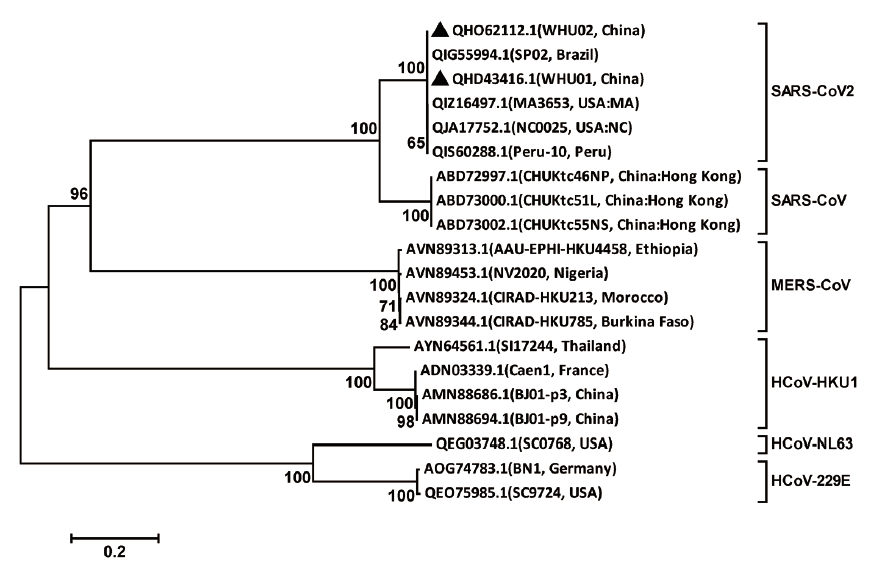
图1 SARS-CoV-2及代表冠状病毒的S蛋白氨基酸序列系统发生树
Fig.1 Phylogenetic tree of amino acid sequence of S protein in SARS-CoV-2 strain and other representative viruses of Coronavirus

图2 SARS-CoV-2 S蛋白与SARS-CoV S蛋白的氨基酸序列比对 红色方框指示目标区段,下同
Fig.2 Sequence alignment of amino acid sequence of S protein in SARS-CoV-2 and SARS-CoV strain Red box indicates selected region, the same below
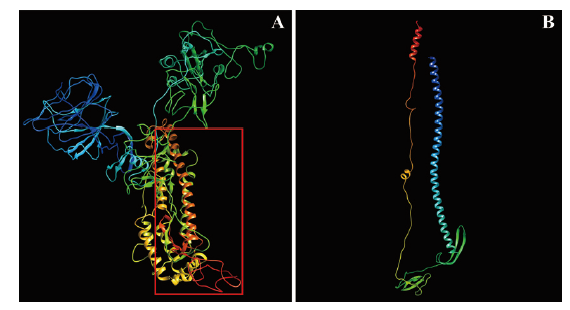
图4 Wuhan-Hu-1株S蛋白整体及目标区段的结构预测 A: S蛋白结构;B: 目标区段局部放大结构
Fig.4 Structure prediction of whole and target region of S protein in Wuhan-Hu-1 strain A: Structure prediction of S protein. B: Local magnification structure of target region
| 参数Parameter | 位置Location |
|---|---|
| 柔韧性 Flexibility(Karplus-Schulz) | 946-950,952-956,964-969,982-987,994,996-1 005,1 008-1 011,1 028-1 030,1 034-1 039,1 045,1 053-1 056,1 070-1 077,1 084-1 086,1 090-1 093,1 097-1 100,1 105-1 108,1 116-1 125,1 135-1 155,1 157-1 164,1 166-1 172,1 180-1 186,1 191-1 196,1 202-1 207 |
| 亲水性 Hydrophilicity(Kyte-Doolittle) | 951,954-955,962,966,983-985,987,992,1 000,1 002,1 004,1 007-1 008,1 011,1 016,1 028,1 036-1 039,1 054-1 056,1 068-1 076,1 085,1 088-1 090,1 099,1 105-1 111,1 117-1 118,1 136-1 137,1 140-1 144,1 146-1 162,1 179-1 186,1 189,1 192-1 193,1 203-1 210 |
| 表面可能性 Surface probability(Emini) | 951,954-955,962,966,983-985,987,992,1 000,1 002,1 004,1 007-1 008,1 011,1 016,1 028,1 036-1 039,1 054-1 056,1 068-1 076,1 085,1 088-1 090,1 099,1 105-1 111,1 117-1 118,1 136-1 137,1 140-1 144,1 146-1 162,1 179-1 186,1 189,1 192-1 193,1 203-1 210 |
| 抗原指数 Antigenic index(Jameson-Wolf) | 944,947-950,952-956,966-967,980-993,995-1 000,1 002-1003,1 011,1 014,1 016-1 022,1 028-1 031,1 035-1 048,1 054-1 058,1 070-1 077,1 082-1 099,1 106- 1 111,1 116-1 119,1 123-1 126,1 136-1 171,1 179-1 199,1 201-1 208 |
表2 Wuhan-Hu-1株S蛋白目标区段的柔韧性、亲水性、表面可能性和抗原指数
Table 2 Flexibility regions, hydrophilicity, surface probability and antigenic index of S protein target region in Wuhan-Hu-1 strain
| 参数Parameter | 位置Location |
|---|---|
| 柔韧性 Flexibility(Karplus-Schulz) | 946-950,952-956,964-969,982-987,994,996-1 005,1 008-1 011,1 028-1 030,1 034-1 039,1 045,1 053-1 056,1 070-1 077,1 084-1 086,1 090-1 093,1 097-1 100,1 105-1 108,1 116-1 125,1 135-1 155,1 157-1 164,1 166-1 172,1 180-1 186,1 191-1 196,1 202-1 207 |
| 亲水性 Hydrophilicity(Kyte-Doolittle) | 951,954-955,962,966,983-985,987,992,1 000,1 002,1 004,1 007-1 008,1 011,1 016,1 028,1 036-1 039,1 054-1 056,1 068-1 076,1 085,1 088-1 090,1 099,1 105-1 111,1 117-1 118,1 136-1 137,1 140-1 144,1 146-1 162,1 179-1 186,1 189,1 192-1 193,1 203-1 210 |
| 表面可能性 Surface probability(Emini) | 951,954-955,962,966,983-985,987,992,1 000,1 002,1 004,1 007-1 008,1 011,1 016,1 028,1 036-1 039,1 054-1 056,1 068-1 076,1 085,1 088-1 090,1 099,1 105-1 111,1 117-1 118,1 136-1 137,1 140-1 144,1 146-1 162,1 179-1 186,1 189,1 192-1 193,1 203-1 210 |
| 抗原指数 Antigenic index(Jameson-Wolf) | 944,947-950,952-956,966-967,980-993,995-1 000,1 002-1003,1 011,1 014,1 016-1 022,1 028-1 031,1 035-1 048,1 054-1 058,1 070-1 077,1 082-1 099,1 106- 1 111,1 116-1 119,1 123-1 126,1 136-1 171,1 179-1 199,1 201-1 208 |
| 区段 Sequence number | 起始位置 Initial position | 终止位置 Terminal position | B细胞抗原表位氨基酸序列 Amino acid sequence of B cell antigenic epitope | 序列长度 Length/aa |
|---|---|---|---|---|
| 1 | 959 | 966 | LNTLVKQL | 8 |
| 2 | 973 | 979 | ISSVLND | 7 |
| 3 | 1 003 | 1 011 | SLQTYVTQQ | 9 |
| 4 | 1 030 | 1 037 | SECVLGQS | 8 |
| 5 | 1 057 | 1 070 | PHGVVFLHVTYVPA | 14 |
| 6 | 1 079 | 1 085 | PAICHDG | 7 |
| 7 | 1 123 | 1 132 | SGNCDVVIGI | 10 |
| 8 | 1 174 | 1 179 | ASVVNI | 6 |
表3 可与SARS-CoV发生交叉免疫反应的SARS-CoV-2株S蛋白B细胞抗原表位
Table 3 B cell epitopes of S protein in SARS-CoV-2 strain which can cross-immunize with SARS-CoV
| 区段 Sequence number | 起始位置 Initial position | 终止位置 Terminal position | B细胞抗原表位氨基酸序列 Amino acid sequence of B cell antigenic epitope | 序列长度 Length/aa |
|---|---|---|---|---|
| 1 | 959 | 966 | LNTLVKQL | 8 |
| 2 | 973 | 979 | ISSVLND | 7 |
| 3 | 1 003 | 1 011 | SLQTYVTQQ | 9 |
| 4 | 1 030 | 1 037 | SECVLGQS | 8 |
| 5 | 1 057 | 1 070 | PHGVVFLHVTYVPA | 14 |
| 6 | 1 079 | 1 085 | PAICHDG | 7 |
| 7 | 1 123 | 1 132 | SGNCDVVIGI | 10 |
| 8 | 1 174 | 1 179 | ASVVNI | 6 |
| [1] |
Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China[J]. Lancet, 2020, 395(10223):497-506.
doi: 10.1016/S0140-6736(20)30183-5 URL |
| [2] |
Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern[J]. Lancet, 2020, 395(10223):470-473.
doi: 10.1016/S0140-6736(20)30185-9 URL |
| [3] | 中国疾病预防控制中心. 新型冠状病毒肺炎流行病学特征分析[J]. 中华流行病学杂志, 2020(2):145-151. |
| Chinese Center for Disease Control and Prevention. Epidemiological analysis of novel coronavirus pneumonia[J]. Chinese Journal of Epidemiology, 2020(2):145-151. | |
| [4] |
Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019[J]. N Engl J Med, 2020, 382(8):727-733.
doi: 10.1056/NEJMoa2001017 URL |
| [5] |
Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation[J]. Science, 2020, 367(6483):1260-1263.
doi: 10.1126/science.abb2507 URL |
| [6] |
Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2[J]. Science, 2020, 367(6485):1444-1448.
doi: 10.1126/science.abb2762 URL |
| [7] |
Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin[J]. Nature, 2020, 579(7798):270-273.
doi: 10.1038/s41586-020-2012-7 URL |
| [8] |
Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus:implications for virus origins and receptor binding[J]. Lancet, 2020, 395(10224):565-574.
doi: 10.1016/S0140-6736(20)30251-8 URL |
| [9] |
Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor[J]. Nature, 2020, 581(7807):215-220.
doi: 10.1038/s41586-020-2180-5 URL |
| [10] |
Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China[J]. Nature, 2020, 579(7798):265-269.
doi: 10.1038/s41586-020-2008-3 URL |
| [11] |
Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody[J]. Nature, 2020, 583:290-295.
doi: 10.1038/s41586-020-2349-y URL |
| [12] |
Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein[J]. Cell, 2020, 181(2):281-292.
doi: 10.1016/j.cell.2020.02.058 URL |
| [13] |
Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors[J]. Science, 2020, 368(6489):409-412.
doi: 10.1126/science.abb3405 URL |
| [14] | 朱锡华, 吴玉章. 对表位生物学研究的认识和体会[J]. 上海免疫学杂志, 1998(1):1-2. |
| Zhu XH, Wu YH. Understanding and experience of epitope biology studies[J]. Shanghai Journal of Immunology, 1998(1):1-2. | |
| [15] |
Krueger DK, Kelly SM, Lewicki DN, et al. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion[J]. J Virol, 2001, 75(6):2792-2802.
pmid: 11222703 |
| [16] |
Walls AC, Tortorici MA, Snijder J, et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion[J]. PNAS, 2017, 114(42):11157-11162.
doi: 10.1073/pnas.1708727114 URL |
| [17] |
Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus(SARS-CoV-2)based on SARS-CoV immunological studies[J]. Viruses, 2020, 12(3):254.
doi: 10.3390/v12030254 URL |
| [18] |
Mcroy WC, Baric RS. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion[J]. J Virol, 2008, 82(3):1414-1424.
doi: 10.1128/JVI.01674-07 URL |
| [19] | 沈媚, 陈冰清, 于瑞嵩, 等. 冠状病毒S蛋白及其受体的结构和功能[J]. 微生物学通报, 2017, 44(10):2452-2462. |
| Shen M, Chen BQ, Yu RS, et al. Structure and function of coronavirus S proteins and their receptors[J]. Microbiology Bulletin, 2017, 44(10):2452-2462. | |
| [20] | 毛亚萍, 卞大伟. SARS-CoV-2毒株S蛋白突变及其影响的生物信息学分析[J]. 病毒学报, 2020, 36(6):1020-1027. |
| Mao YP, Bian DW. Bioinformatics analysis of S protein in SARS-CoV-2 strains and their effects[J]. Journal of Viruses, 2020, 36(6):1020-1027. | |
| [21] |
Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 Spike Glycoprotein[J]. Cell, 2020, 181(2):281-292.
doi: 10.1016/j.cell.2020.02.058 URL |
| [22] | Xia S, Yan L, Xu W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike[J]. Sci Adv, 2019, 5(4):v4580. |
| [23] |
Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences[J]. PNAS, 1981, 78(6):3824-3828.
pmid: 6167991 |
| [24] |
Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein[J]. J Mol Biol, 1982, 157(1):105-132.
pmid: 7108955 |
| [25] | 孙建宏, 曹殿军. 细胞的抗原表位研究方法[J]. 动物医学进展, 2004, 25(5):18-21. |
| Sun JH, Cao DJ. An antigenic epitope study of cells[J]. Advances in Animal Medicine, 2004, 25(5):18-21. | |
| [26] |
Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses[J]. Trends Microbiol, 2016, 24(6):490-502.
doi: 10.1016/j.tim.2016.03.003 URL |
| [27] |
Liu L, Wei Q, Lin Q, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection[J]. JCI Insight, 2019, 4(4):e123158.
doi: 10.1172/jci.insight.123158 URL |
| [28] | 周建平, 周裕军, 许秀丽, 等. 北京丹大生物技术有限公司.新型冠状病毒优势表位融合蛋白、诊断试剂及应用制造技术:中国, 20942233[P]. 2020-06-02. |
| Zhou JP, Zhou YJ, Xu XL, et al. Novel Coronavirus dominant epitope fusion protein, diagnostic reagent and applied manufacturing technology:China, 20942233[P]. 2020-06-02. |
| [1] | 陈晓萌, 张雪静, 张欢, 张宝江, 苏艳. 重组牛乳源金黄色葡萄球菌GapC蛋白优势B细胞抗原表位的预测和筛选[J]. 生物技术通报, 2023, 39(5): 306-313. |
| [2] | 汪巧菊, 胡雨萌, 温亚亚, 宋丽, 孟闯, 潘志明, 焦新安. 新型冠状病毒S1蛋白的表达及活性鉴定[J]. 生物技术通报, 2022, 38(3): 157-163. |
| [3] | 陈铎, 刘永哲. 新型冠状病毒核蛋白N端结构域的表达纯化以及晶体优化[J]. 生物技术通报, 2022, 38(12): 149-155. |
| [4] | 张西西, 张怡青, 李玉林, 韩笑, 王国强, 王晓军, 王旭东, 王云龙. 新型冠状病毒(SARS-CoV-2)N蛋白C端重组蛋白的原核表达、纯化及应用[J]. 生物技术通报, 2021, 37(5): 92-97. |
| [5] | 陆盼盼, 郭松林, 关瑞章, 冯建军. 致病性弧菌外膜蛋白及其免疫原性研究进展[J]. 生物技术通报, 2014, 0(4): 30-35. |
| [6] | 王慧, 罗成波, 贺纛, 孔令萍, 周碧君, 文明, 程振涛, 王开功. 绵羊肺炎支原体DnaK B细胞抗原表位预测及其蛋白结构分析[J]. 生物技术通报, 2014, 0(3): 159-164. |
| [7] | 赵文娟;田亮;薄新文;马勋;康立超;. 牦牛多杀性巴氏杆菌外膜蛋白OmpH的扩增与生物信息学分析[J]. , 2012, 0(01): 139-144. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||