








生物技术通报 ›› 2023, Vol. 39 ›› Issue (6): 286-297.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1303
陈宝强( ), 李莹莹, 马博雅, 肉扎古丽·马利克, 优丽图孜·乃比, 宋金迪, 刘君(
), 李莹莹, 马博雅, 肉扎古丽·马利克, 优丽图孜·乃比, 宋金迪, 刘君( ), 王希东(
), 王希东( )
)
收稿日期:2022-10-25
出版日期:2023-06-26
发布日期:2023-07-07
通讯作者:
刘君,女,博士,副教授,研究方向:植物病原细菌学;E-mail: liujem@126.com;作者简介:陈宝强,男,硕士,研究方向:植物病原细菌与植物互作机制;E-mail: q1144757863@126.com
基金资助:
CHEN Bao-qiang( ), LI Ying-ying, MA Bo-ya, ROUZHAGULI Malike, YOULITUZI Naibi, SONG Jin-di, LIU Jun(
), LI Ying-ying, MA Bo-ya, ROUZHAGULI Malike, YOULITUZI Naibi, SONG Jin-di, LIU Jun( ), WANG Xi-dong(
), WANG Xi-dong( )
)
Received:2022-10-25
Published:2023-06-26
Online:2023-07-07
摘要:
III型分泌效应物(type III secreted effectors, T3SEs)是细菌性果斑病(bacterial fruit blotch, BFB)的病原菌——西瓜食酸菌(Acidovorax citrulli)分泌的关键致病因子。鉴定西瓜食酸菌特异的、具有GNAT(Gcn5-related N-acetyltransferase)超家族结构域的T3SE基因aop2,分析其编码蛋白质影响植物免疫的方式,可为深入认识该基因在病菌致病机制中的作用奠定基础。利用生物信息学分析其序列特征;借助荧光定量PCR技术分析aop2的表达调控及其表达与抗病相关基因表达间的关系;利用基因突变及基因功能互补手段,通过分析致病性、寄主活性氧积累量等解析基因功能;使用瞬时表达技术了解Aop2抑制激发子诱导的细胞坏死能力及其亚细胞定位情况。aop2基因启动子区存在III型分泌系统(type III secretion system, T3SS)核心基因结合位点,其编码的蛋白不存在信号肽和跨膜螺旋区,含一个GNAT家族乙酰转移酶结构域但无同源蛋白;T3SS核心基因hrpG/hrpX突变体中aop2基因的表达量显著降低;缺失aop2基因的突变体对寄主黄瓜的致病力降低,但黄瓜子叶中活性氧积累量增加;Aop2可定位于烟草整个细胞,能够抑制由坏死因子NIP诱导的PCD(programmed cell death);Aop2的表达增强了烟草叶片中病原相关分子模式触发的免疫(PAMP-triggered immunity, PTI)信号通路,以及SA和JA信号通路相关基因的表达。结果表明,Aop2为西瓜食酸菌一个含有GNAT结构域的特异T3SE,其在与寄主黄瓜互作中发挥毒性因子功能,在与烟草互作中参与调控植物细胞死亡及PTI和植物激素相关抗病防卫反应。
陈宝强, 李莹莹, 马博雅, 肉扎古丽·马利克, 优丽图孜·乃比, 宋金迪, 刘君, 王希东. 西瓜食酸菌III型分泌效应物基因aop2功能分析[J]. 生物技术通报, 2023, 39(6): 286-297.
CHEN Bao-qiang, LI Ying-ying, MA Bo-ya, ROUZHAGULI Malike, YOULITUZI Naibi, SONG Jin-di, LIU Jun, WANG Xi-dong. Functional Analysis of the Type III Secreted Effector Gene aop2 in Acidovorax citrulli[J]. Biotechnology Bulletin, 2023, 39(6): 286-297.
| 菌株和质粒 Strain and plasmid | 名称 Name | 特性 Characteristic | 来源Source |
|---|---|---|---|
| 菌株 Strain | FC440(WT) | AmpR; wild type | Our laboratory |
| FC440(∆aop2) | AmpR; KanR; FC440 mutant defective in aop2 | This study | |
| FC440(∆aop2-aop2) | AmpR; KanR; GmR; FC440(∆aop2)complemented with the fragment of aop2 gene expressed by vector pBBR1MCS-5 | This study | |
| FC440(∆hrpG) | AmpR; KanR; FC440 mutant defective in hrpG | [14] | |
| FC440(∆hrpX) | AmpR; KanR; FC440 mutant defective in hrpX | [14] | |
| Trans T1(pBBR1MCS-5- aop2) | GmR; E. coli TransT1 strain containing vector pBBR1MCS-5-aop2 | This study | |
| S17-1(pBBR1MCS-5- aop2) | GmR; E. coli S17-1 strain containing vector pBBR1MCS-5aop2 | This study | |
| GV3101 | RifR; wild type | Our laboratory | |
| GV3101(pAPK-aop2) | SpecR; RifR; GmR; GV3101 strain containing vector pAPK-aop2 | This study | |
| GV3101(pAPK-GFP) | SpecR; RifR; GmR; GV3101 strain containing vector pAPK-GFP | [15] | |
| GV3101(pBINGFP2-aop2) | KanR; RifR; GmR; GV3101 strain containing vector pBINGFP2-aop2 | This study | |
| GV3101(pBINGFP2) | SpecR; RifR; GmR; GV3101 strain containing vector pBINGFP2 | [15] | |
| GV3101(BAX) | KanR; RifR; GmR; GV3101 strain containing BAX | [15] | |
| GV3101(NIP) | KanR; RifR; GmR; GV3101 strain containing NEP | [15] | |
| GV3101(NIP) | KanR; RifR; GmR; GV3101 strain containing NEP | [15] | |
| GV3101(Avh241) | KanR; RifR; GmR; GV3101 strain containing Avh241 | [15] | |
| GV3101(INF1) | KanR; RifR; GmR; GV3101 strain containing INF1 | [15] | |
| 质粒 Plasmid | pMD19-T | AmpR; Cloning vector | TaKaRa |
| pMD19-T- aop2 | AmpR; pMD19-T vector containing a 438-bp fragment with the aop2 gene | This study | |
| pK19mob2ΩHMB-aop2 | KanR; pK19mob2ΩHMB vector containing a 301-bp fragment with the aop2 gene | This study | |
| pBBR1MCS-5-aop2 | GmR; pBBR1MCS-5 vector containing aop2 gene; used to complement FC440(∆aop2) | This study | |
| pKannibal-aop2 | KanR; pKannibal vector containing a 438-bp fragment with the aop2 gene | This study | |
| pAPK-aop2 | SpecR; pART27 vevtor containing pKannibal-aop2 | This study | |
| pBINGFP2-aop2 | KanR; pBINGFP2 vector containing a 438-bp fragment with the aop2 gene | This study |
表1 供试菌株、质粒
Table 1 Bacterial strains, plasmids used in this study
| 菌株和质粒 Strain and plasmid | 名称 Name | 特性 Characteristic | 来源Source |
|---|---|---|---|
| 菌株 Strain | FC440(WT) | AmpR; wild type | Our laboratory |
| FC440(∆aop2) | AmpR; KanR; FC440 mutant defective in aop2 | This study | |
| FC440(∆aop2-aop2) | AmpR; KanR; GmR; FC440(∆aop2)complemented with the fragment of aop2 gene expressed by vector pBBR1MCS-5 | This study | |
| FC440(∆hrpG) | AmpR; KanR; FC440 mutant defective in hrpG | [14] | |
| FC440(∆hrpX) | AmpR; KanR; FC440 mutant defective in hrpX | [14] | |
| Trans T1(pBBR1MCS-5- aop2) | GmR; E. coli TransT1 strain containing vector pBBR1MCS-5-aop2 | This study | |
| S17-1(pBBR1MCS-5- aop2) | GmR; E. coli S17-1 strain containing vector pBBR1MCS-5aop2 | This study | |
| GV3101 | RifR; wild type | Our laboratory | |
| GV3101(pAPK-aop2) | SpecR; RifR; GmR; GV3101 strain containing vector pAPK-aop2 | This study | |
| GV3101(pAPK-GFP) | SpecR; RifR; GmR; GV3101 strain containing vector pAPK-GFP | [15] | |
| GV3101(pBINGFP2-aop2) | KanR; RifR; GmR; GV3101 strain containing vector pBINGFP2-aop2 | This study | |
| GV3101(pBINGFP2) | SpecR; RifR; GmR; GV3101 strain containing vector pBINGFP2 | [15] | |
| GV3101(BAX) | KanR; RifR; GmR; GV3101 strain containing BAX | [15] | |
| GV3101(NIP) | KanR; RifR; GmR; GV3101 strain containing NEP | [15] | |
| GV3101(NIP) | KanR; RifR; GmR; GV3101 strain containing NEP | [15] | |
| GV3101(Avh241) | KanR; RifR; GmR; GV3101 strain containing Avh241 | [15] | |
| GV3101(INF1) | KanR; RifR; GmR; GV3101 strain containing INF1 | [15] | |
| 质粒 Plasmid | pMD19-T | AmpR; Cloning vector | TaKaRa |
| pMD19-T- aop2 | AmpR; pMD19-T vector containing a 438-bp fragment with the aop2 gene | This study | |
| pK19mob2ΩHMB-aop2 | KanR; pK19mob2ΩHMB vector containing a 301-bp fragment with the aop2 gene | This study | |
| pBBR1MCS-5-aop2 | GmR; pBBR1MCS-5 vector containing aop2 gene; used to complement FC440(∆aop2) | This study | |
| pKannibal-aop2 | KanR; pKannibal vector containing a 438-bp fragment with the aop2 gene | This study | |
| pAPK-aop2 | SpecR; pART27 vevtor containing pKannibal-aop2 | This study | |
| pBINGFP2-aop2 | KanR; pBINGFP2 vector containing a 438-bp fragment with the aop2 gene | This study |
| Primer name | Sequnece(5'-3') | Source |
|---|---|---|
| aop2-F | CGGAATTCTGCTGGAGGTCCTGCTGT(EcoR I) | This study |
| aop2-R | CCAAGCTTACTTCCATCGCATCGTCC(Hind III) | This study |
| aop2-QF | CCCTCGAGTGGCAACGATGTTTCCCC(Xho I) | This study |
| aop2-QR | GCTCTAGAGCGACAGCGACGGCTGAG(Xba I) | This study |
| AAC-1 | GACCAGCCCACAACTGGGAC | [16] |
| AAC-2 | CTGCCGCACTCCAGCGA | [16] |
| aop2-CF | TTTGGAGAGGACACGCTCGAGATGACAGACAGACTCAGCCGGC(Xho I) | This study |
| aop2-CR | TCATTAAAGCAGGACTCTAGATCAGTTCACCGTTGACGACGC(Xba I) | This study |
| aop2-DF | AGAGGATCCGTCGACCCCGGGATGAGCCTGCAGAACGCCA(Sma I) | This study |
| aop2-DR | CTGTACAAGGGTACCCCCGGGTCAGGCGTCCAGGGGCAG(Sma I) | This study |
| RT-aop2-F | CCTGGCTGGCGGACAAGT | This study |
| RT-aop2-R | CTGCCCGAAGAACTGCGA | This study |
| rpoB-F | GCGACAGCGTGCTCAAAGTG | [17] |
| rpoB-R | GGCCTTCGTTGGTGCGTTTCT | [17] |
| NbPti5-F | CCTCCAAGTTTGAGCTCGGATAGT | [18] |
| NbPti5-R | CCAAGAAATTCTCCATGCACTCTGTC | [18] |
| NbAcre31-F | AATTCGGCCATCGTGATCTTGGTC | [18] |
| NbAcre31-R | GAGAAACTGGGATTGCCTGAAGGA | [18] |
| NbGras2-F | TACCTAGCACCAAGCAGATGCAGA | [18] |
| NbGras2-R | TCATGAGGCGTTACTCGGAGCATT | [18] |
| NbEF1α-F | AAGGTCCAGTATGCCTGGGTGCTTGAC | [18] |
| NbEF1α-R | AAGAATTCACAGGGAC AGTTCCAATACCA | [18] |
| NbWRKY7-F | CACAAGGGTACAAACAACACAG | [19] |
| NbWRKY7-R | GGTTGCATTTGGTTCATGTAAG | [19] |
| NbWRKY8-F | AACAATGGTGCCAATAATGC | [19] |
| NbWRKY8-R | TGCATATCCTGAGAAACCATT | [19] |
| NbPR2b-F | TCCAACTTGGAATCAAAGGG | [20] |
| NbPR2b-R | GTGGACACTATACTCAGGTG | [20] |
| NbLOX-F | AAAACCTATGCCTCAAGAAC | [20] |
| NbLOX-R | ACTGCTGCATAGGCTTTGG | [20] |
| NbEFR1-F | GCTCTTAACGTCGGATGGTC | [20] |
| NbEFR1-R | AGCCAAACCCTAGCTCCATT | [20] |
| NbGAPDH-F | AGCTCAAGGGAATTCTCGATG | [21] |
| NbGAPDH-R | AACCTTAACCATGTCATCTCCC | [21] |
表2 供试引物
Table 2 Primers used in this study
| Primer name | Sequnece(5'-3') | Source |
|---|---|---|
| aop2-F | CGGAATTCTGCTGGAGGTCCTGCTGT(EcoR I) | This study |
| aop2-R | CCAAGCTTACTTCCATCGCATCGTCC(Hind III) | This study |
| aop2-QF | CCCTCGAGTGGCAACGATGTTTCCCC(Xho I) | This study |
| aop2-QR | GCTCTAGAGCGACAGCGACGGCTGAG(Xba I) | This study |
| AAC-1 | GACCAGCCCACAACTGGGAC | [16] |
| AAC-2 | CTGCCGCACTCCAGCGA | [16] |
| aop2-CF | TTTGGAGAGGACACGCTCGAGATGACAGACAGACTCAGCCGGC(Xho I) | This study |
| aop2-CR | TCATTAAAGCAGGACTCTAGATCAGTTCACCGTTGACGACGC(Xba I) | This study |
| aop2-DF | AGAGGATCCGTCGACCCCGGGATGAGCCTGCAGAACGCCA(Sma I) | This study |
| aop2-DR | CTGTACAAGGGTACCCCCGGGTCAGGCGTCCAGGGGCAG(Sma I) | This study |
| RT-aop2-F | CCTGGCTGGCGGACAAGT | This study |
| RT-aop2-R | CTGCCCGAAGAACTGCGA | This study |
| rpoB-F | GCGACAGCGTGCTCAAAGTG | [17] |
| rpoB-R | GGCCTTCGTTGGTGCGTTTCT | [17] |
| NbPti5-F | CCTCCAAGTTTGAGCTCGGATAGT | [18] |
| NbPti5-R | CCAAGAAATTCTCCATGCACTCTGTC | [18] |
| NbAcre31-F | AATTCGGCCATCGTGATCTTGGTC | [18] |
| NbAcre31-R | GAGAAACTGGGATTGCCTGAAGGA | [18] |
| NbGras2-F | TACCTAGCACCAAGCAGATGCAGA | [18] |
| NbGras2-R | TCATGAGGCGTTACTCGGAGCATT | [18] |
| NbEF1α-F | AAGGTCCAGTATGCCTGGGTGCTTGAC | [18] |
| NbEF1α-R | AAGAATTCACAGGGAC AGTTCCAATACCA | [18] |
| NbWRKY7-F | CACAAGGGTACAAACAACACAG | [19] |
| NbWRKY7-R | GGTTGCATTTGGTTCATGTAAG | [19] |
| NbWRKY8-F | AACAATGGTGCCAATAATGC | [19] |
| NbWRKY8-R | TGCATATCCTGAGAAACCATT | [19] |
| NbPR2b-F | TCCAACTTGGAATCAAAGGG | [20] |
| NbPR2b-R | GTGGACACTATACTCAGGTG | [20] |
| NbLOX-F | AAAACCTATGCCTCAAGAAC | [20] |
| NbLOX-R | ACTGCTGCATAGGCTTTGG | [20] |
| NbEFR1-F | GCTCTTAACGTCGGATGGTC | [20] |
| NbEFR1-R | AGCCAAACCCTAGCTCCATT | [20] |
| NbGAPDH-F | AGCTCAAGGGAATTCTCGATG | [21] |
| NbGAPDH-R | AACCTTAACCATGTCATCTCCC | [21] |

图2 Aop2序列的生物信息学分析 A:aop2基因转录起始位点上游PIP-box分析(第一行为革兰氏阴性菌T3SEs启动子区的保守基序的模体,第二行为aop2基因启动子区序列,A、T、C、G、B、N、Y、R为碱基单字母缩写);B:Aop2蛋白的信号肽预测;C:Aop2蛋白的跨膜螺旋区预测;D:Aop2蛋白的结构域预测
Fig. 2 Bioinformatics analysis of Aop2 A: PIP-box analysis existed upstream of aop2 gene transcription start site(The first line is the motifs of the conservative sequences in the T3SEs promoter region of Gram-negative bacteria. The second line is the sequence of the aop2 gene promoter region. A, T, C, G, B, N, Y, and R are the single letter abbreviations of bases). B: Signal peptide prediction of Aop2 protein. C: Transmembrane spiral region prediction of Aop2 protein. D: Domain prediction of Aop2 protein
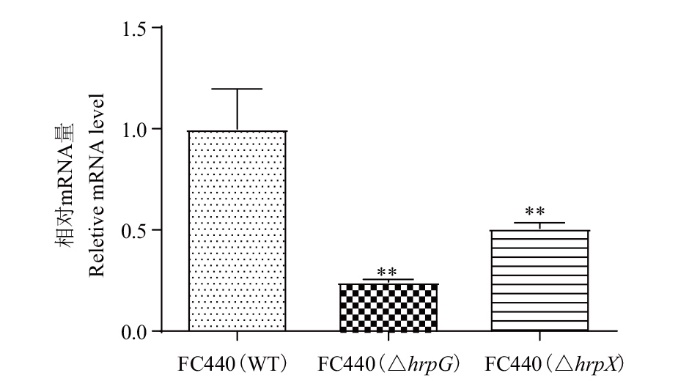
图3 aop2基因在西瓜食酸菌hrpG/hrpX基因突变体中的表达分析 **表示在0.01水平上存在差异。下同
Fig. 3 Expression analysis of aop2 gene in hrpG or hrpX gene mutant in A. citrulli ** indicate significant difference at the 0.01 probability. The same below

图4 西瓜食酸菌aop2基因突变体及其衍生菌株的致病力表型 A:黄瓜子叶发病表型(图片采集于接菌1 d时);B:病情指数分析;C:秦烟95的HR表型(图片采集于接菌1 d时)
Fig. 4 Virulence phenotype of aop2 mutants and their derived strains of A. citrulli A: The phenotype of cucumber cotyledon(Photos were taken at 1 d after inoculation). B: Disease index analysis. C: HR phenotype of Qinyan 95(Photos were taken at 1 d after inoculation)
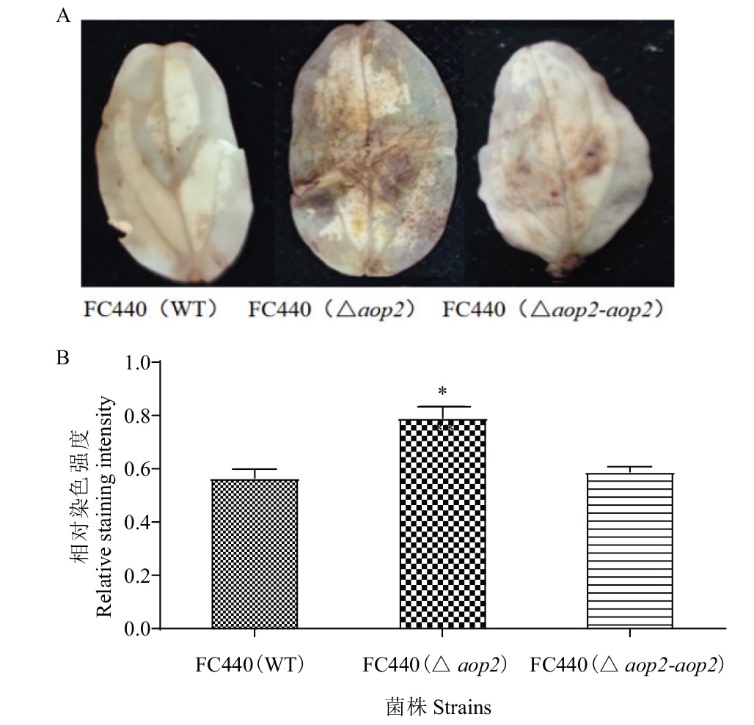
图5 西瓜食酸菌aop2基因突变体及其衍生菌株侵染下黄瓜ROS产量分析 A:黄瓜子叶ROS染色表型(图片采集于叶片接菌后1 d时);B:相对染色强度分析。*表示在0.05平上存在差异
Fig. 5 Analysis of ROS production of cucumber infected by aop2 mutants and and their derived strains of A. citrulli A: ROS staining phenotype of cucumber cotyledon (Photos were taken at 1 d after inoculation). B: Relative staining intensity analysis. * indicates significant difference at the 0.05 probability
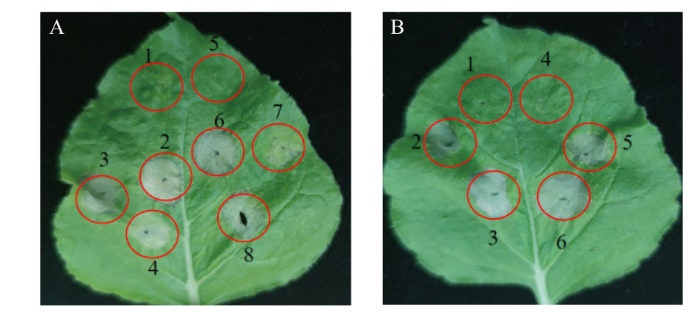
图7 Aop2对5种激发子诱导PCD的抑制作用
Fig. 7 Inhibitory effect of Aop2 on the PCD induced by five elicitors A: Inhibitory effect of Aop2 on BAX, NIP and Avh241; 1: GV3101(pAPK-GFP); 2: GV3101(pAPK-GFP)+GV3101(BAX); 3: GV3101(pAPK-GFP)+GV3101(NIP); 4: GV3101(pAPK-GFP)+GV3101(Avh241); 5: GV3101(pAPK-aop2); 6: GV3101(pAPK-aop2)+GV3101(BAX); 7: GV3101(pAPK-aop2)+GV3101(NIP); 8: GV3101(pAPK-aop2)+GV3101(Avh241). B: Inhibitory effect of Aop2 on BAX, NIP and Avh241; 1: GV3101(pAPK-GFP); 2: GV3101(pAPK-GFP)+GV3101(NEP); 3: GV3101(pAPK-GFP)+GV3101(INF1); 4: GV3101(pAPK-aop2); 5: GV3101(pAPK-aop2)+GV3101(NEP); 6: GV3101(pAPK-aop2)+GV3101(INF1)
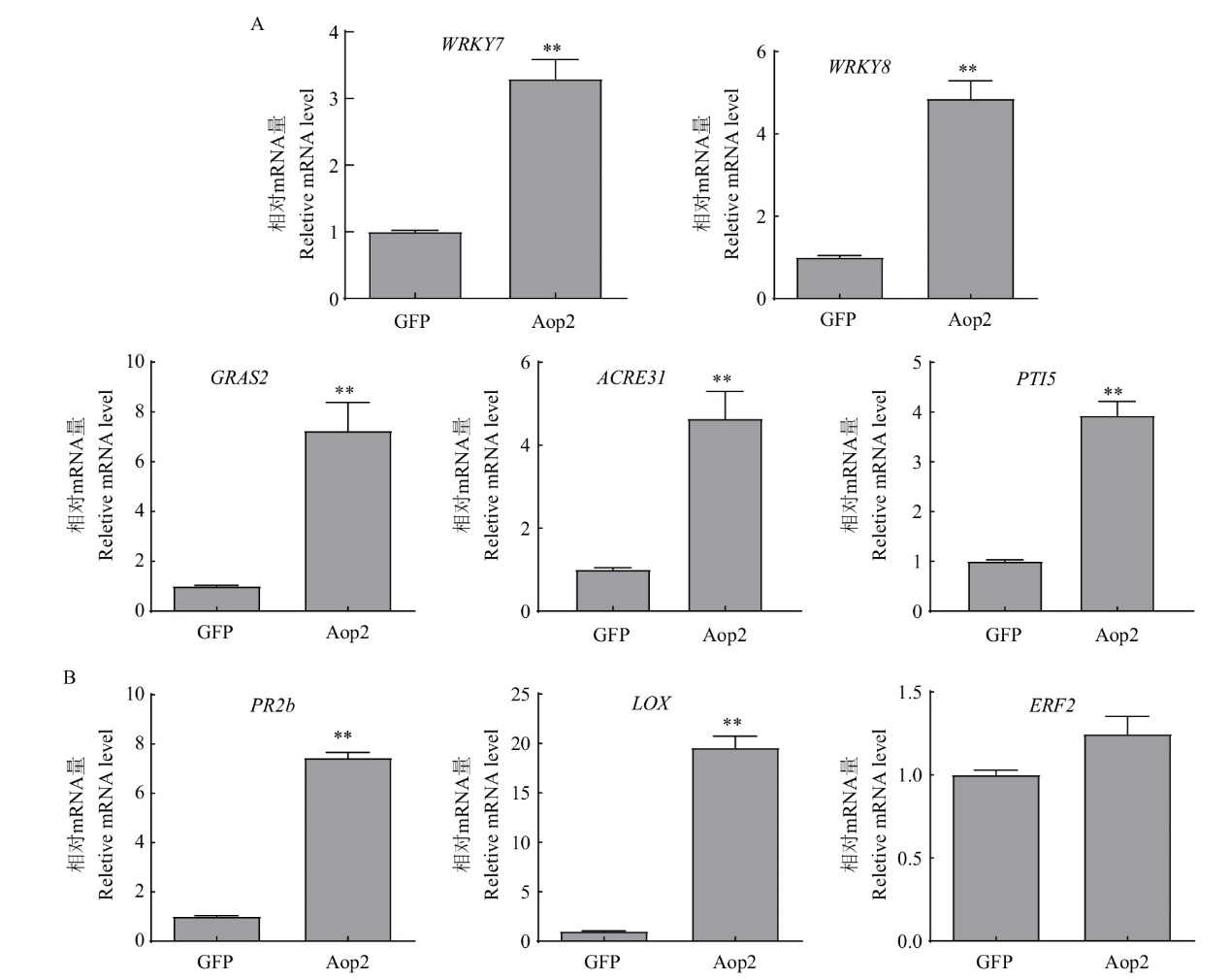
图8 Aop2对本氏烟抗病信号通路的影响 A:Aop2对PTI信号通路的影响分析;B:Aop2对植物激素信号通路的影响分析(叶片于瞬时表达36 h时取样)
Fig. 8 Effects of Aop2 on the resistance signal pathway in N. benthamiana A: Analysis of the effect of Aop2 on PTI signal pathway. B: Analysis of the effect of Aop2 on plant hormone signal pathway(Leaf samples were taken 36 h after transient expression)
| [1] |
Burdman S, Walcott R. Acidovorax citrulli: generating basic and applied knowledge to tackle a global threat to the cucurbit industry[J]. Mol Plant Pathol, 2012, 13(8): 805-815.
doi: 10.1111/j.1364-3703.2012.00810.x pmid: 22738439 |
| [2] |
Ge YX, Luo L, Xia LM, et al. Fermentation: an unreliable seed treatment for bacterial fruit blotch of watermelon[J]. Plant Dis, 2021, 105(4): 1026-1033.
doi: 10.1094/PDIS-05-20-1056-RE URL |
| [3] |
Kumagai LB, Woods PW, Walcott R, et al. First report of bacterial fruit blotch on melon caused by Acidovorax citrulli in California[J]. Plant Dis, 2014, 98(10): 1423.
doi: 10.1094/PDIS-03-14-0286-PDN pmid: 30704008 |
| [4] |
Chung MY, Kim H, Beuchat LR, et al. Antimicrobial activities of plant essential oil vapours against Acidovorax citrulli and Xanthomonas campestris on Cucurbitaceae, Brassicaceae and Solanaceae seeds[J]. J Appl Microbiol, 2022, 132(3): 2189-2202.
doi: 10.1111/jam.15352 URL |
| [5] |
Bahar O, de la Fuente L, Burdman S. Assessing adhesion, biofilm formation and motility of Acidovorax citrulli using microfluidic flow Chambers[J]. FEMS Microbiol Lett, 2010, 312(1): 33-39.
doi: 10.1111/j.1574-6968.2010.02094.x pmid: 20807236 |
| [6] |
Zhang XX, Zhao M, Yan JP, et al. Involvement of hrpX and hrpG in the virulence of Acidovorax citrulli strain Aac5, causal agent of bacterial fruit blotch in cucurbits[J]. Front Microbiol, 2018, 9: 507.
doi: 10.3389/fmicb.2018.00507 URL |
| [7] |
Eckshtain-Levi N, Munitz T, Živanović M, et al. Comparative analysis of type III secreted effector genes reflects divergence of Acidovorax citrulli strains into three distinct lineages[J]. Phytopathology, 2014, 104(11): 1152-1162.
doi: 10.1094/PHYTO-12-13-0350-R pmid: 24848275 |
| [8] |
Jiménez-Guerrero I, Pérez-Montaño F, da Silva GM, et al. Show me your secret(ed)weapons: a multifaceted approach reveals a wide arsenal of type III-secreted effectors in the cucurbit pathogenic bacterium Acidovorax citrulli and novel effectors in the Acidovorax genus[J]. Mol Plant Pathol, 2020, 21(1): 17-37.
doi: 10.1111/mpp.12877 pmid: 31643123 |
| [9] |
Zhang XX, Zhao M, Jiang J, et al. Identification and functional analysis of AopN, an Acidovorax citrulli effector that induces programmed cell death in plants[J]. Int J Mol Sci, 2020, 21(17): 6050.
doi: 10.3390/ijms21176050 URL |
| [10] | 张晓晓. 西瓜噬酸菌效应蛋白Ace1功能研究及光照黑暗条件下致病性差异分析[D]. 北京: 中国农业科学院, 2018. |
| Zhang XX. Functional study of effector Ace1 & analysis of pathogenicity differences under light and dark conditions in Acidovorax citrulli[D]. Beijing: Chinese Academy of Agricultural Sciences, 2018. | |
| [11] | 杨琳琳. 西瓜噬酸菌效应蛋白Ace0201和Ace1242的鉴定及生物学功能初步分析[D]. 沈阳: 沈阳农业大学, 2019. |
| Yang LL. Identification and biological function of the effector Ace0201 and Ace1242 in Acidovorax citrulli[D]. Shenyang: Shenyang Agricultural University, 2019. | |
| [12] |
Kong L, Qiu XF, Kang JG, et al. A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection[J]. Curr Biol, 2017, 27(7): 981-991.
doi: 10.1016/j.cub.2017.02.044 URL |
| [13] | 马畅, 李婷, 刘士尧, 等. 蛋白乙酰化酶GNAT调控水稻细菌性条斑病菌的致病力[J]. 海南大学学报: 自然科学版, 2019, 37(1): 29-33. |
| Ma C, Li T, Liu SY, et al. GNAT regulates the virulence of Xanthomonas oryzae pv. Oryzicola[J]. Nat Sci J Hainan Univ, 2019, 37(1): 29-33. | |
| [14] | 优丽图孜·乃比. 西瓜食酸菌与黄瓜互作转录组分析及T3SEs基因的初步鉴定[D]. 乌鲁木齐: 新疆农业大学, 2021. |
| Youlituzi N. Transcriptome analysis of Acidovorax citrulli—cucumber interaction and preliminary identification of T3SE genes in Acidovorax citrulli[D]. Urumqi: Xinjiang Agricultural University, 2021. | |
| [15] | 张强. 寄生疫霉菌胞外分泌效应蛋白Pp18和PpCys44/45的功能分析[D]. 杨凌: 西北农林科技大学, 2019. |
| Zhang Q. Functional analysis of the effector proteins Pp18 and PpCys44/45 secreted by Phytophthora parasitica[D]. Yangling: Northwest A & F University, 2019. | |
| [16] |
Walcott RR, Fessehaie A, Castro AC. Differences in pathogenicity between two genetically distinct groups of Acidovorax avenae subsp. citrulli on cucurbit hosts[J]. J Phytopathol, 2004, 152(5): 277-285.
doi: 10.1111/j.1439-0434.2004.00841.x URL |
| [17] | 优丽图孜·乃比, 王希东, 刘君, 等. 西瓜食酸菌与黄瓜互作转录组分析[J]. 微生物学通报, 2021, 48(10): 3667-3681. |
| Youlituzi N, Wang XD, Liu J, et al. Transcriptome analysis of the interaction between Acidovorax citrulli and cucumber[J]. Microbiol China, 2021, 48(10): 3667-3681. | |
| [18] |
Chen CL, Liu SS, Liu Q, et al. An ANNEXIN-like protein from the cereal cyst nematode Heterodera avenae suppresses plant defense[J]. PLoS One, 2015, 10(4): e0122256.
doi: 10.1371/journal.pone.0122256 URL |
| [19] | 刘琳硕, 贺祥, 李红梅, 等. TRV介导的E3泛素连接酶基因NbE3R14沉默对烟草基础免疫及南方根结线虫寄生的影响[J]. 南京农业大学学报, 2020, 43(1): 65-71. |
| Liu LS, He X, Li HM, et al. Effects of TRV-mediated silencing of the E3 ubiquitin ligase gene NbE3R14 in Nicotiana benthamiana on the plant basal immunity and the parasitism of Meloidogyne incognita[J]. J Nanjing Agric Univ, 2020, 43(1): 65-71. | |
| [20] | 张美祥, 安玉艳, 刘廷利, 等. 在本氏烟中瞬时表达效应因子PsCRN127基因提高其对寄生疫霉的抗性[J]. 南京农业大学学报, 2015, 38(6): 930-935. |
| Zhang MX, An YY, Liu TL, et al. Transient expression of the Ps- CRN127 effector gene enhances Nicotiana benthamiana resistance to Phytophthora parasitica[J]. J Nanjing Agric Univ, 2015, 38(6): 930-935. | |
| [21] | 池俊玲, 赵一博, 郭江波, 等. 不同浓度Cd2+胁迫下烟草实时荧光定量PCR内参基因的筛选[J]. 南方农业学报, 2019, 50(10): 2133-2140. |
| Chi JL, Zhao YB, Guo JB, et al. Screening of internal reference genes for real-time fluorescence quantitative PCR under different concentrations of Cd2+ stress in tobacco[J]. J South Agric, 2019, 50(10): 2133-2140. | |
| [22] | 颉兵兵, 刘君, 优丽图孜·乃比, 等. 西瓜食酸菌抗铜基因cueR的生物信息学分析及功能验证[J]. 微生物学通报, 2020, 47(5): 1534-1543. |
| Xie BB, Liu J, Youlituzi N, et al. Bioinformatics analysis and functional verification of copper resistance gene cueR in Acidovorax citrulli[J]. Microbiol China, 2020, 47(5): 1534-1543. | |
| [23] |
Shirakawa T. Studies on control of seed borne bacterial vegetable disease: studies on epidemiology and control of cucurbits bacterial fruit blotch[J]. J Gen Plant Pathol, 2021, 87(6): 408-412.
doi: 10.1007/s10327-021-01034-5 |
| [24] | Fan QR, Bibi S, Vallad GE, et al. Identification of genes in Xanthomonas euvesicatoria pv. rosa that are host limiting in tomato[J]. Plants(Basel), 2022, 11(6): 796. |
| [25] |
Arnold R, Brandmaier S, Kleine F, et al. Sequence-based prediction of type III secreted proteins[J]. PLoS Pathog, 2009, 5(4): e1000376.
doi: 10.1371/journal.ppat.1000376 URL |
| [26] |
Zhang XX, Yang YW, Zhao M, et al. Acidovorax citrulli type III effector AopP suppresses plant immunity by targeting the watermelon transcription factor WRKY6[J]. Front Plant Sci, 2020, 11: 579218.
doi: 10.3389/fpls.2020.579218 URL |
| [27] |
Wei HL, Chakravarthy S, Mathieu J, et al. Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB[J]. Cell Host Microbe, 2015, 17(6): 752-762.
doi: 10.1016/j.chom.2015.05.007 URL |
| [28] |
Zhang MX, Li Q, Liu TL, et al. Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases[J]. Plant Physiol, 2015, 167(1): 164-175.
doi: 10.1104/pp.114.252437 URL |
| [29] | 张美祥, 刘廷利, 茹艳艳, 等. 效应因子PsCRN77基因在本氏烟中的表达降低其对寄生疫霉的抗性[J]. 植物病理学报, 2015, 45(6): 619-625. |
| Zhang MX, Liu TL, Ru YY, et al. Expression of an effector gene PsCRN77 decreases Nicotiana benthamiana resistance to oomycete pathogen Phytophthora parasitica[J]. Acta Phytopathol Sin, 2015, 45(6): 619-625. | |
| [30] |
Lewis JD, Wilton M, Mott GA, et al. Immunomodulation by the Pseudomonas syringae HopZ type III effector family in Arabidopsis[J]. PLoS One, 2014, 9(12): e116152.
doi: 10.1371/journal.pone.0116152 URL |
| [31] |
Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes[J]. Annu Rev Phytopathol, 2006, 44: 41-60.
pmid: 16448329 |
| [32] | 龚玄云静. 组蛋白乙酰转移酶Gcn5间接调控基因表达的分子机制[D]. 武汉: 湖北大学, 2020. |
| Gong XYJ. Exploration of mechanism of indirect gene regulation by histone acetyltransferase Gcn5[D]. Wuhan: Hubei University, 2020. | |
| [33] |
Kim S, Piquerez SJM, Ramirez-Prado JS, et al. GCN5 modulates salicylic acid homeostasis by regulating H3K14ac levels at the 5' and 3' ends of its target genes[J]. Nucleic Acids Res, 2020, 48(11): 5953-5966.
doi: 10.1093/nar/gkaa369 pmid: 32396165 |
| [34] |
Guéneron M, Timmers AC, Boucher C, et al. Two novel proteins, PopB, which has functional nuclear localization signals, and PopC, which has a large leucine-rich repeat domain, are secreted through the hrp-secretion apparatus of Ralstonia solanacearum[J]. Mol Microbiol, 2000, 36(2): 261-277.
pmid: 10792715 |
| [35] |
Ma KW, Jiang SS, Hawara E, et al. Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor[J]. New Phytol, 2015, 208(4): 1157-1168.
doi: 10.1111/nph.2015.208.issue-4 URL |
| [36] | Aung K, Xin XF, Mecey C, et al. Subcellular localization of Pseudomonas syringae pv. tomato effector proteins in plants[J]. Methods Mol Biol, 2017, 1531: 141-153. |
| [37] |
Göhre V, Spallek T, Häweker H, et al. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB[J]. Curr Biol, 2008, 18(23): 1824-1832.
doi: 10.1016/j.cub.2008.10.063 pmid: 19062288 |
| [38] |
Li LX, Atef A, Piatek A, et al. Characterization and DNA-binding specificities of Ralstonia TAL-like effectors[J]. Mol Plant, 2013, 6(4): 1318-1330.
doi: 10.1093/mp/sst006 URL |
| [39] |
Li GY, Froehlich JE, Elowsky C, et al. Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts[J]. Plant J, 2014, 77(2): 310-321.
doi: 10.1111/tpj.2013.77.issue-2 URL |
| [40] | Traore S. Characterization of type three effector genes of A. citrulli, the causal agent of bacterial fruit blotch of cucurbits[D]. Blacksburg: Virginia Polytechnic Institute and State University, 2014. |
| [1] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [2] | 苏雨, 李宗芸, 韩永华. 植物液泡加工酶研究进展[J]. 生物技术通报, 2021, 37(6): 181-191. |
| [3] | 李凯晴, 李莹, 王艺磊, 邹鹏飞. 受体相互作用蛋白的功能及在硬骨鱼类中的研究进展[J]. 生物技术通报, 2021, 37(5): 197-211. |
| [4] | 查星琴, 杨明华, 李永能, 赵素梅, 黄英. Leptin介导JAK/STAT信号通路对猪皮下前脂肪细胞脂类代谢调控的研究[J]. 生物技术通报, 2021, 37(2): 88-95. |
| [5] | 胡濒月, 胡杨, 成文敏, 赵素梅, 赵红业, 魏红江. Leptin过表达对猪前脂肪细胞脂滴形成的研究[J]. 生物技术通报, 2020, 36(8): 111-119. |
| [6] | 刘辉, 邓治, 杨洪, 代龙军, 李德军. 橡胶树HbMC2在酵母中的表达和抗逆性分析[J]. 生物技术通报, 2018, 34(9): 202-208. |
| [7] | 朱海峰, 吴丹, 吴敬. 毕赤酵母GS115中N-乙酰转移酶在大肠杆菌中的克隆表达与性质研究[J]. 生物技术通报, 2015, 31(11): 179-185. |
| [8] | 李辉,卢守亮,万鹏程,代蓉,李良远,孟浩浩,石国庆. Leptin基因多态性与绵羊季节性发情的相关分析[J]. 生物技术通报, 2014, 0(9): 189-194. |
| [9] | 程抒劼;黄仕杰;郭丽琼;韩飞;林俊芳;. 南方红豆杉10-去乙酰巴卡亭Ⅲ-10-乙酰转移酶基因的克隆与生物信息学分析[J]. , 2011, 0(01): 107-112. |
| [10] | 赵建平;姚祥坦;. 植物小孢子胚胎发生机理[J]. , 2009, 0(06): 7-11. |
| [11] | 赵强;赵志文;张廷婷;崔德才;王斌. 豇豆胰蛋白酶抑制剂(CpTI)基因转化欧美杨的研究[J]. , 2005, 0(04): 54-58. |
| [12] | 李凤娜;陈晓安. 一氧化氮对线粒体生物合成的积极影响[J]. , 2005, 0(02): 57-57. |
| [13] | 李凤娜;陈晓安. 一氧化氮对线粒体生物合成的积极影响[J]. , 2005, 0(02): 57-57. |
| [14] | 秦春圃. 转基因作物及其产品的检测[J]. , 2003, 0(04): 50-51. |
| [15] | . 国外动态[J]. , 2000, 0(02): 41-46. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||