








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 68-79.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0248
邓嘉辉1( ), 雷建峰2, 赵燚1, 刘敏1, 胡子曜1, 尤扬子1, 邵武奎1, 柳建飞1, 刘晓东1(
), 雷建峰2, 赵燚1, 刘敏1, 胡子曜1, 尤扬子1, 邵武奎1, 柳建飞1, 刘晓东1( )
)
收稿日期:2023-03-21
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
刘晓东,男,博士,教授,研究方向:作物逆境分子生物学;E-mail: xiaodongliu75@aliyun.com作者简介:邓嘉辉,男,硕士,研究方向:作物逆境分子生物学;E-mail: 15754842489@126.com
基金资助:
DENG Jia-hui1( ), LEI Jian-feng2, ZHAO Yi1, LIU Min1, HU Zi-yao1, YOU Yang-zi1, SHAO Wu-kui1, LIU Jian-fei1, LIU Xiao-dong1(
), LEI Jian-feng2, ZHAO Yi1, LIU Min1, HU Zi-yao1, YOU Yang-zi1, SHAO Wu-kui1, LIU Jian-fei1, LIU Xiao-dong1( )
)
Received:2023-03-21
Published:2023-10-26
Online:2023-11-28
摘要:
MCP是MS2噬菌体的外壳蛋白,Csy4是一种参与CRISPR 1-F系统crRNA生成的小型蛋白,能够以较高的特异性识别并结合RNA。目前CRISPR/Cas等基因组编辑技术存在靶向核酸酶分子量大、脱靶率高、受PAM位点限制等问题,为解决上述问题,构建基于上述两种小型蛋白的新型迷你基因组编辑系统。本研究采用AlphaFold2预测MCP-FokI、FokI-MCP、Csy4-FokI和FokI-Csy4融合蛋白的结构,通过浸花法将MCP-FokI和FokI-MCP编辑载体分别转化拟南芥,利用拟南芥叶片注射的方法投送CLCrV介导的Csy4-FokI与FokI-Csy4编辑系统,提取拟南芥基因组DNA,通过HI-TOM高通量测序检测新型迷你基因组编辑系统的编辑能力。结果显示,融合蛋白中MCP、FokI和Csy4都各自保持着自身原有的三维结构,预示它们都能正常发挥彼此的功能。构建靶向敲除拟南芥CLA1基因的4个不同中间间隔区的双靶位点MCP-FokI和MCP-FokI植物表达载体,初步证明MCP-FokI和FokI-MCP均不能实现对靶基因的靶向编辑。构建靶向敲除拟南芥CLA1基因的7个CLCrV介导的不同中间间隔区的双靶位点Csy4-FokI编辑载体,其中CLCrV介导的Csy4-FokI编辑系统能够实现对靶基因的靶向编辑,但是突变类型均为碱基置换类型且编辑效率很低,而FokI-Csy4基因组编辑体系并未检测到编辑的发生。成功构建了Csy4-FokI新型迷你基因组编辑系统,为克服CRISPR/Cas基因组编辑技术存在的问题提供了一种新的解决方案。
邓嘉辉, 雷建峰, 赵燚, 刘敏, 胡子曜, 尤扬子, 邵武奎, 柳建飞, 刘晓东. 基于Csy4与MCP的新型迷你基因组编辑系统的构建[J]. 生物技术通报, 2023, 39(10): 68-79.
DENG Jia-hui, LEI Jian-feng, ZHAO Yi, LIU Min, HU Zi-yao, YOU Yang-zi, SHAO Wu-kui, LIU Jian-fei, LIU Xiao-dong. Construction of a New Mini Genome Editing System Based on Csy4 and MCP[J]. Biotechnology Bulletin, 2023, 39(10): 68-79.
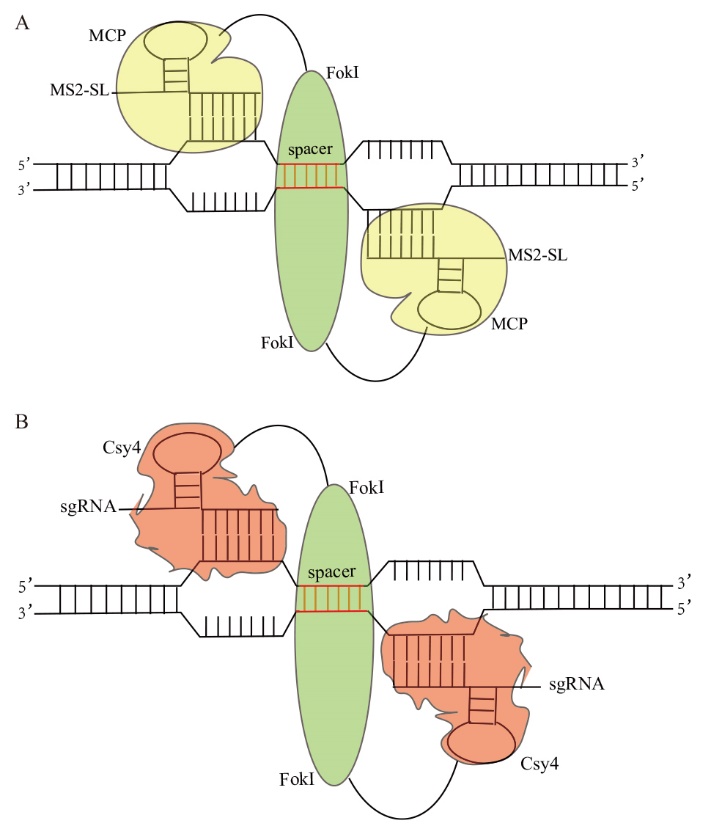
图1 MCP-FokI(FokI-MCP)和Csy4-FokI(FokI-Csy4)基因组编辑体系示意图 A:MS2-SL引导的FokI核酸酶原理概述:两个MCP-FokI融合蛋白招募两个不同的MS2-SL靶向到目标基因位点上,促进Fok I切割双链DNA断裂;B:sgRNA引导的FokI核酸酶原理概述:两个Csy4-FokI融合蛋白招募两个不同的sgRNA靶向到目标基因位点上,促进Fok I切割双链DNA断裂
Fig. 1 Schematic diagram of MCP-FokI(FokI-MCP)and Csy4-FokI(FokI-Csy4)genome editing system A: Overview of MS2-SL-guided FokI nuclease principle: Two MCP-FokI fusion proteins recruit two different MS2-SLs to the target gene locus, promoting Fok I cleavage of the double-stranded DNA. B: Overview of sgRNA-guided FokI nuclease principle: Two Csy4-FokI fusion proteins recruit two different sgRNAs to the target gene locus, promoting Fok I cleavage of the double-stranded DNA
| 引物Primer | 序列Sequence(5'-3') | 用途Application |
|---|---|---|
| FokI-Link-Csy4-F | CCTCCGGGGGATCTTCAGGAGGATCAATGGGAGATCATTATCTTGATATTAG | 构建FokI-Csy4 Construction of FokI-Csy4 |
| FokI-Link-Csy4-R | GCCTTTTTCGTGGCCGCCGGCCTTTTGAACCAAGGAACAAAACCTC | |
| T-Csy4-FokI-F | GGTCGGTATCCACGGAGTCCCAGCAGCCATGGGAGATCATTATCTTGATATTAG | 构建Csy4-FokI Construction of Csy4-FokI |
| T-Csy4-FokI-R | GACGATCCGCCGGAGGACCCTCCAGAGAACCAAGGAACAAAACCTC | |
| T-sgRNA1-F | TAGAGTCGACATAGCGATTGGTCATTAAGAATCACAATCGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA1 Construction of AtU6∷sgRNA1 |
| T-sgRNA1-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA2-F | GAGTCGACATAGCGATTGAATCACAATCATATCAGAGGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA2 Construction of AtU6∷sgRNA2 |
| T-sgRNA2-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA3-F | CTAGAGTCGACATAGCGATTGAAGAATCACAATCATATCAGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA3 Construction of AtU6∷sgRNA3 |
| T-sgRNA3-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA4-F | GAGTCGACATAGCGATTGAGAATCACAATCATATCAGGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA4 Construction of AtU6∷sgRNA4 |
| T-sgRNA4-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA5-F | GTCGAAGTAGTGATTGGCTTATGAAGCCATGAACAGTTCACTGCCGTATAGGC | 构建AtU6-26∷sgRNA5 Construction of AtU6-26∷sgRNA5 |
| T-sgRNA5-R | TTGTCGACGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA6-F | GTAGTGATTGGAAGCCATGAACAACGCCGGTTCACTGCCGTATAGGC | 构建AtU6-26∷sgRNA6 Construction of AtU6-26∷sgRNA6 |
| T-sgRNA6-R | TTGTCGACGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| KP-U6-SK-F | GGTACCTTAATTAAGGAGTGATCAAAAGTCCCAC | 构建Kpn I-Pac I-AtU6∷sgRNA(1 2 3 4)-EcoR I Construction of Kpn I-Pac I-AtU6∷sgRNA(1 2 3 4)-EcoR I |
| E-U6-SK-R | GAATTCGATCCGGATCTCGAGTAAGA | |
| E-U6-26-SK-F | GAATTCGAATTGCCCTTAAGCTTCGTTG | 构建EcoR I-AtU6-26∷sgRNA(5 6)-Xba I-Avr II Construction of EcoR I-AtU6-26∷sgRNA(5 6)-Xba I-Avr II |
| XM-U6-26-SK-R | TCTAGACCTAGGGATCCGGATCTCGAGTAAGA | |
| AT-FC/CF-F | GACCACCAACTCCATTACTTG | PCR扩增AtCLA1基因涵盖靶位点区域 The PCR amplification is performed to target and amplify the specific region of the AtCLA1 |
| AT-FC/CF-R | GAGAGCCGGGTTAGACTGTA | |
| Spe I-CLF-1F | ACTAGTTGCCTGCAGGTCAACATG | 构建Spe I-FokI-Csy4(Csy4-FokI)-Pac I Construction of Spe I-FokI-Csy4(Csy4- FokI)-Pac I |
| Pac I-CLF-1r | TTAATTAATAGTAACATAGATGACACCGC | |
| HiT-CLA1-F | GGAGTGAGTACGGTGTGCGGTTGCTGTGATTGGTGATGG | 高通量扩增AtCLA1基因涵盖靶位点区域 High-throughput amplification is performed to cover the target region of the AtCLA1 |
| HiT-CLA1-R | GAGTTGGATGCTGGATGGTCCATCCAAAGTAGCTGTAGGT | |
| CLCrVA-F | GCTGCTTGTATGTTTGGGTG | CLCrV-A 病毒检测 CLCrV-A virus detection |
| CLCrVA-R | CAAGGGAGAGTAGTTGGCA |
表1 引物及用途
Table 1 Primers and applications
| 引物Primer | 序列Sequence(5'-3') | 用途Application |
|---|---|---|
| FokI-Link-Csy4-F | CCTCCGGGGGATCTTCAGGAGGATCAATGGGAGATCATTATCTTGATATTAG | 构建FokI-Csy4 Construction of FokI-Csy4 |
| FokI-Link-Csy4-R | GCCTTTTTCGTGGCCGCCGGCCTTTTGAACCAAGGAACAAAACCTC | |
| T-Csy4-FokI-F | GGTCGGTATCCACGGAGTCCCAGCAGCCATGGGAGATCATTATCTTGATATTAG | 构建Csy4-FokI Construction of Csy4-FokI |
| T-Csy4-FokI-R | GACGATCCGCCGGAGGACCCTCCAGAGAACCAAGGAACAAAACCTC | |
| T-sgRNA1-F | TAGAGTCGACATAGCGATTGGTCATTAAGAATCACAATCGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA1 Construction of AtU6∷sgRNA1 |
| T-sgRNA1-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA2-F | GAGTCGACATAGCGATTGAATCACAATCATATCAGAGGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA2 Construction of AtU6∷sgRNA2 |
| T-sgRNA2-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA3-F | CTAGAGTCGACATAGCGATTGAAGAATCACAATCATATCAGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA3 Construction of AtU6∷sgRNA3 |
| T-sgRNA3-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA4-F | GAGTCGACATAGCGATTGAGAATCACAATCATATCAGGTTCACTGCCGTATAGGC | 构建AtU6∷sgRNA4 Construction of AtU6∷sgRNA4 |
| T-sgRNA4-R | CCTTTCTAGAGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA5-F | GTCGAAGTAGTGATTGGCTTATGAAGCCATGAACAGTTCACTGCCGTATAGGC | 构建AtU6-26∷sgRNA5 Construction of AtU6-26∷sgRNA5 |
| T-sgRNA5-R | TTGTCGACGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| T-sgRNA6-F | GTAGTGATTGGAAGCCATGAACAACGCCGGTTCACTGCCGTATAGGC | 构建AtU6-26∷sgRNA6 Construction of AtU6-26∷sgRNA6 |
| T-sgRNA6-R | TTGTCGACGCCCGGGATCCGGATCTCGAGTAAGAGCTCG | |
| KP-U6-SK-F | GGTACCTTAATTAAGGAGTGATCAAAAGTCCCAC | 构建Kpn I-Pac I-AtU6∷sgRNA(1 2 3 4)-EcoR I Construction of Kpn I-Pac I-AtU6∷sgRNA(1 2 3 4)-EcoR I |
| E-U6-SK-R | GAATTCGATCCGGATCTCGAGTAAGA | |
| E-U6-26-SK-F | GAATTCGAATTGCCCTTAAGCTTCGTTG | 构建EcoR I-AtU6-26∷sgRNA(5 6)-Xba I-Avr II Construction of EcoR I-AtU6-26∷sgRNA(5 6)-Xba I-Avr II |
| XM-U6-26-SK-R | TCTAGACCTAGGGATCCGGATCTCGAGTAAGA | |
| AT-FC/CF-F | GACCACCAACTCCATTACTTG | PCR扩增AtCLA1基因涵盖靶位点区域 The PCR amplification is performed to target and amplify the specific region of the AtCLA1 |
| AT-FC/CF-R | GAGAGCCGGGTTAGACTGTA | |
| Spe I-CLF-1F | ACTAGTTGCCTGCAGGTCAACATG | 构建Spe I-FokI-Csy4(Csy4-FokI)-Pac I Construction of Spe I-FokI-Csy4(Csy4- FokI)-Pac I |
| Pac I-CLF-1r | TTAATTAATAGTAACATAGATGACACCGC | |
| HiT-CLA1-F | GGAGTGAGTACGGTGTGCGGTTGCTGTGATTGGTGATGG | 高通量扩增AtCLA1基因涵盖靶位点区域 High-throughput amplification is performed to cover the target region of the AtCLA1 |
| HiT-CLA1-R | GAGTTGGATGCTGGATGGTCCATCCAAAGTAGCTGTAGGT | |
| CLCrVA-F | GCTGCTTGTATGTTTGGGTG | CLCrV-A 病毒检测 CLCrV-A virus detection |
| CLCrVA-R | CAAGGGAGAGTAGTTGGCA |

图2 融合蛋白三级结构预测 A:FokI-MCP与MCP-FokI融合蛋白结构预测图;B:FokI-Csy4与Csy4-FokI融合蛋白结构预测图
Fig. 2 Tertiary structure prediction of fusion protein A: Prediction diagram of FokI-MCP and MCP-FokI fusion protein structure.B: Structure prediction of FokI-Csy4 and Csy4-FokI fusion protein

图3 MCP-FokI和FokI-MCP基因编辑载体构建图 M:2 K Plus II DNA标准分子量;WT:野生型;A:以拟南芥AtCLA1为靶标,对于同一个靶点分别用AtU6-26和AtU6启动子转录左侧guide-SL和右侧guide-SL;B:MCP-FokI和FokI-MCP编辑载体构建示意图;C:MCP-FokI和FokI-MCP编辑载体酶切鉴定结果;D:4个不同中间间隔区靶序列过渡载体酶切鉴定结果;E:酶切鉴定(1-8:AtU6-26∷SL1-35∷MCP-FokI-Ter-AtU6∷SL1-P1300, AtU6-26∷SL2-35∷MCP-FokI- Ter-AtU6∷SL2-P1300, AtU6-26∷SL3-35∷MCP-FokI-Ter-AtU6∷SL3-P1300, AtU6-26∷SL4-35∷MCP-FokI-Ter-AtU6∷SL4-P1300, AtU6-26∷SL1-35∷FokI- MCP-Ter-AtU6∷SL1- P1300, AtU6-26∷SL2-35∷FokI-MCP-Ter-AtU6∷SL2-P1300)
Fig. 3 Construction of MCP-FokI and FokI-MCP gene editing vector M: 2 K Plus II DNA standard molecular weight. WT: Wild type. A: AtCLA1 as target, left side-SL and right side-SL transcribed with AtU6-26 and AtU6 promoters respectively for the same target. B: Schematic diagram of MCP-FokI and FokI-MCP editing vector construction. C: MCP-FokI and FokI-MCP editing vector enzymatic identification. D: Enzymatic identification of four different intermediate spacer target sequences. E: Enzymatic identification

图4 MCP-FokI和FokI-MCP基因编辑系统靶向敲除AtCLA1 红色箭头表示转基因抗性苗
Fig. 4 Targeted knockout of AtCLA1 gene by MCP-FokI and FokI-MCP gene editing system The red arrow indicates transgenic resistant seedlings
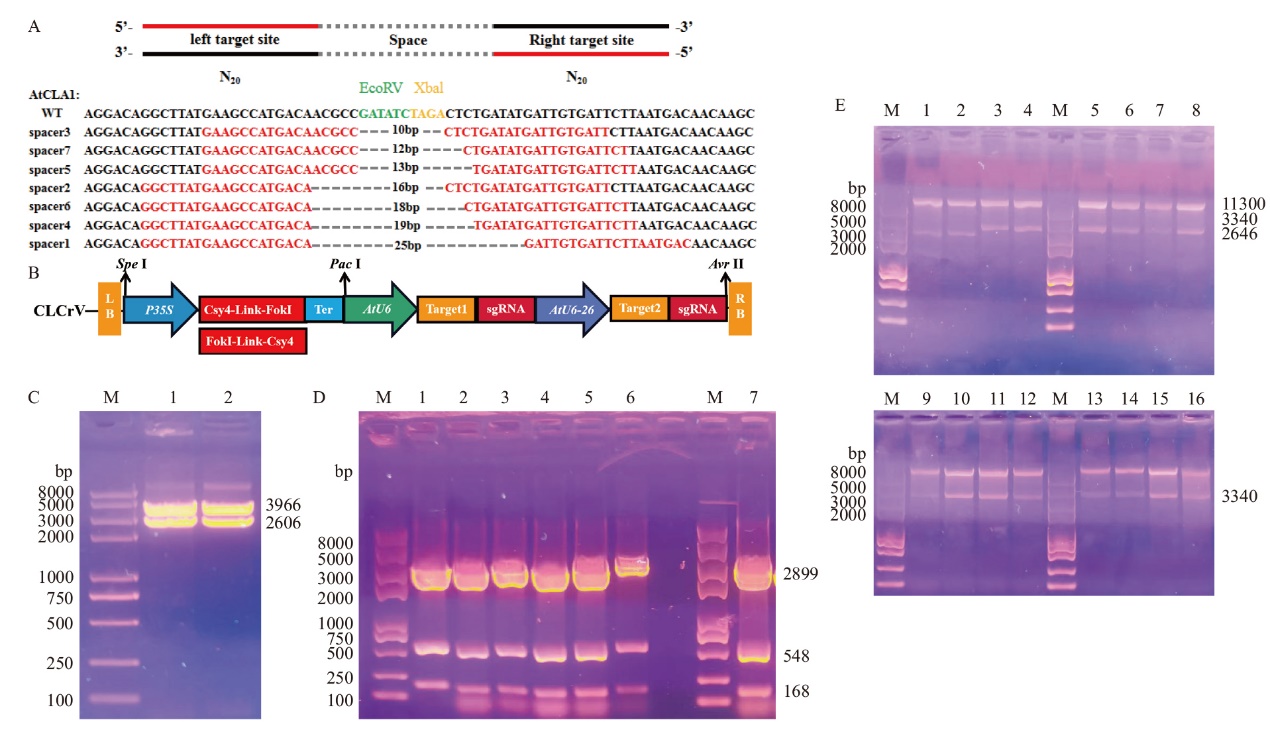
图5 Csy4-FokI和FokI-Csy4基因编辑载体构建图 M:2K Plus II DNA标准分子量;WT:野生型;A:以拟南芥AtCLA1为靶标,对于同一个靶点分别用AtU6-26和AtU6启动子转录左侧sgRNA和右侧sgRNA;B:Csy4-FokI和Csy4-FokI编辑载体构建示意图;C:Csy4-FokI-Bzro和FokI-Csy4-B zro编辑载体酶切鉴定结果;D:7个不同中间间隔区靶序列过渡载体酶切鉴定结果;E:酶切鉴定(1-16:FokI-Csy4-CLCrV、Csy4-FokI-CLCrV, FokI-Csy4-sgRNA(1-5)-CLCrV、FokI-Csy4-sgRNA(2-5)-CLCrV, FokI-Csy4-sgRNA(2-6)-CLCrV, FokI-Csy4-sgRNA(3-5)-CLCrV, FokI-Csy4-sgRNA(3-6)-CLCrV, FokI-Csy4-sgRNA(4-5)-CLCrV, FokI-Csy4-sgRNA(4-6)- CLCrV, Csy4-FokI-sgRNA(1-5)-CLCrV, Csy4-FokI-sgRNA(2-6)-CLCrV, Csy4-FokI-sgRNA(2-6)-CLCrV, Csy4-FokI-sgRNA(3-5)-CLCrV, Csy4-FokI- sgRNA(3-6)-CLCrV, Csy4-FokI-sgRNA(4-5)-CLCrV, Csy4-FokI-sgRNA(4-6)-CLCrV)
Fig. 5 Construction of Csy4-FokI and FokI-Csy4 gene editing vector M: Standard molecular weight of 2K Plus II DNA. WT: Wild type. A: Transcription of left sgRNA and right sgRNA with AtU6-26 and AtU6 promoters respectively for the same target site using AtCLA1 as the target. B: Schematic diagram of Csy4-FokI and Csy4-FokI editing vector construction. C: Csy4-FokI- Bzro and FokI-Csy4-B zro editing vector enzymatic identification results. D: Enzymatic identification results of seven different intermediate spacer region target sequence transition vectors. E: Enzymatic identification
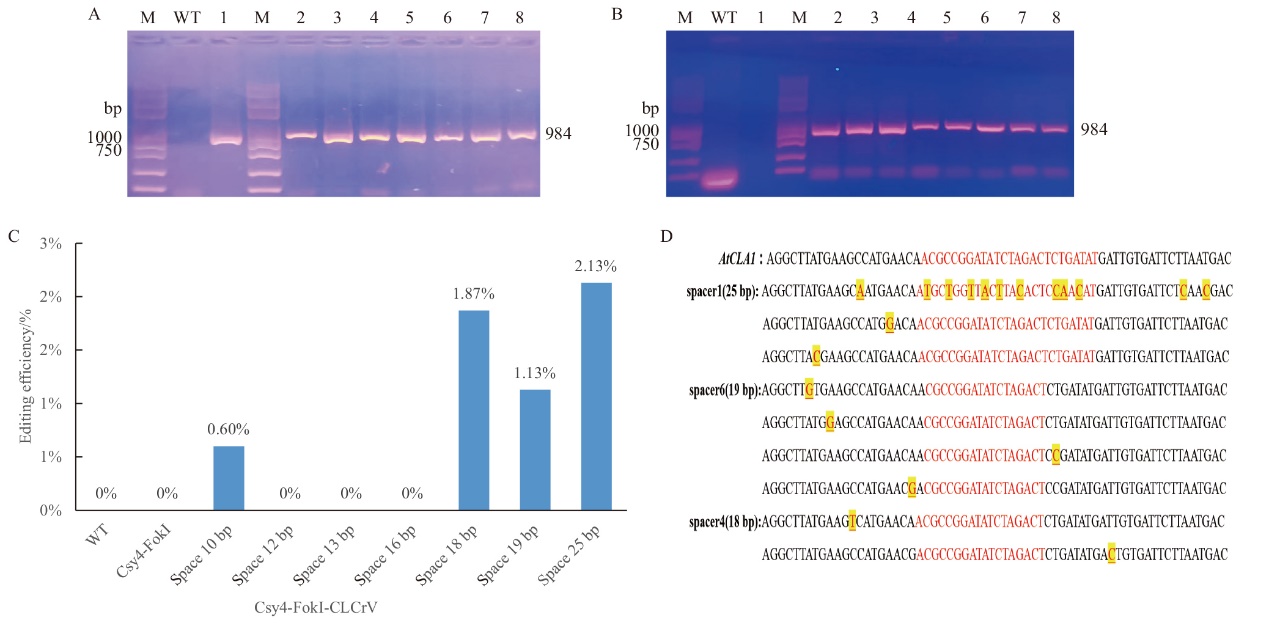
图6 Csy4-FokI和FokI-Csy4基因编辑系统靶向敲除AtCLA1 M:2 K Plus II DNA 标准分子量;A:Csy4-FokI编辑载体成功转入拟南芥后CLCrV-A病毒检测结果(1-8:Csy4-FokI-CLCrV、Csy4-FokI-sgRNA(1-5)-CLCrV、Csy4-FokI-sgRNA(2-6)-CLCrV、Csy4-FokI-sgRNA(2-6)-CLCrV、Csy4-FokI-sgRNA(3-5)-CLCrV、Csy4-FokI-sgRNA(3-6)- CLCrV、Csy4-FokI-sgRNA(4-5)-CLCrV、Csy4-FokI-sgRNA(4-6)-CLCrV);B:FokI-Csy4编辑载体成功转入拟南芥后CLCrV-A病毒检测结果(1-8:FokI-Csy4-CLCrV、FokI-Csy4sgRNA(1-5)-CLCrV、FokI-Csy4-sgRNA(2-6)-CLCrV、FokI-Csy4-sgRNA(2-6)-CLCrV、FokI-Csy4-sgRNA(3-5)-CLCrV、FokI-Csy4-sgRNA(3-6)-CLCrV、FokI-Csy4-sgRNA(4-5)-CLCrV、FokI-Csy4-sgRNA(4-6)-CLCrV);C:CLCrV介导的Csy4-FokI体系对AtCLAI进行编辑的高通量检测;D:瞬时转化Csy4-FokI基因组编辑体系HI-TOM高通量测序分析后得到的突变类型
Fig. 6 Targeted knockout of AtCLA1 gene by Csy4-FokI and FokI-Csy4 gene editing system M: 2 K PlusII DNA standard molecular weight. A: Results of CLCrV-A virus assay after successful transfer of Csy4-FokI editing vector into Arabidopsis thaliana. B: Results of CLCrV-A virus assays after successful transfer of the FokI-Csy4 editing vector into Arabidopsis thaliana. C: High-throughput detection of AtCLAI editing by the CLCrV-mediated Csy4-FokI system. D: Mutation types obtained after high-throughput sequencing analysis of the transiently transformed Csy4-FokI genome editing system HI-TOM
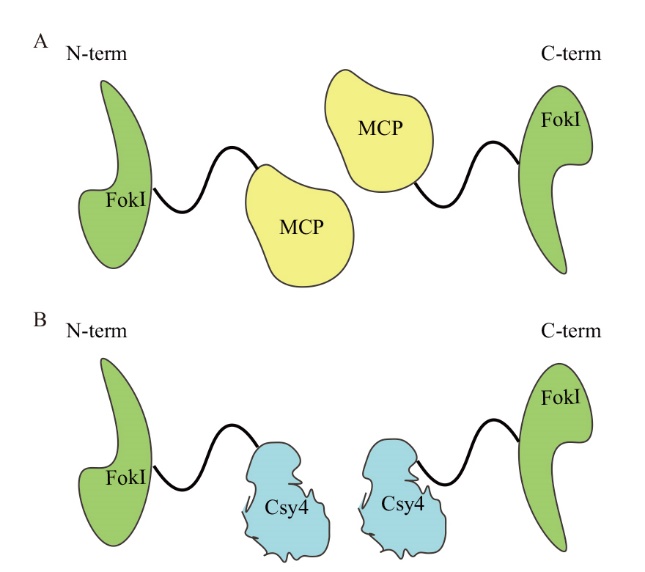
图7 MCP-FokI(FokI-MCP)和Csy4-FokI(FokI-Csy4)融合蛋白示意图 A:MCP-FokI和FokI-MCP融合蛋白,FokI分别位于MCP的N端和C端;B:Csy4-FokI和FokI-Csy4融合蛋白,FokI分别位于Csy4的N端和C端
Fig. 7 Schematic diagram of MCP-FokI(FokI-MCP)and Csy4-FokI(FokI-Csy4)fusion protein A: MCP-FokI and FokI-MCP fusion proteins, FokI are located at the N and C ends of MCP respectively; B: Csy4-FokI and FokI-Csy4 fusion protein, FokI is located at the N terminal and C terminal of Csy4 respectively
| [1] | Zhang YL, Ma XL, Xie XR, et al. CRISPR/Cas9-based genome editing in plants[M]//Progress in Molecular Biology and Translational Science. Amsterdam: Elsevier, 2017: 133-150. |
| [2] |
Mali P, Yang LH, Esvelt KM, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826.
doi: 10.1126/science.1232033 pmid: 23287722 |
| [3] |
Bilichak A, Sastry-Dent L, Sriram S, et al. Genome editing in wheat microspores and haploid embryos mediated by delivery of ZFN proteins and cell-penetrating peptide complexes[J]. Plant Biotechnol J, 2020, 18(5): 1307-1316.
doi: 10.1111/pbi.13296 pmid: 31729822 |
| [4] |
Wilson KA, McEwen AE, Pruett-Miller SM, et al. Expanding the repertoire of target sites for zinc finger nuclease-mediated genome modification[J]. Mol Ther Nucleic Acids, 2013, 2(4): e88.
doi: 10.1038/mtna.2013.13 URL |
| [5] | 康细林, 储丹丹, 单斌. 基因编辑新技术CRISPR-Cas系统研究及应用进展[J]. 国外医药: 抗生素分册, 2020, 41(1): 35-41. |
| Kang XL, Chu DD, Shan B. Progress in research and application of new gene editing technology CRISPR-cas system[J]. World Notes Antibiot, 2020, 41(1): 35-41. | |
| [6] |
Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation[J]. Annu Rev Biochem, 2010, 79: 213-231.
doi: 10.1146/annurev-biochem-010909-095056 pmid: 20192761 |
| [7] |
Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting[J]. Science, 2011, 333(6051): 1843-1846.
doi: 10.1126/science.1204094 pmid: 21960622 |
| [8] |
Char SN, Unger-Wallace E, Frame B, et al. Heritable site-specific mutagenesis using TALENs in maize[J]. Plant Biotechnol J, 2015, 13(7): 1002-1010.
doi: 10.1111/pbi.12344 pmid: 25644697 |
| [9] | 刘蓓, 尉玮, 王丽华. 基因编辑新技术研究进展[J]. 亚热带农业研究, 2013, 9(4): 262-269. |
| Liu B, Yu W, Wang LH. Progress in new techniques of gene editing[J]. Subtrop Agric Res, 2013, 9(4): 262-269. | |
| [10] |
Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nat Biotechnol, 2020, 38(7): 824-844.
doi: 10.1038/s41587-020-0561-9 pmid: 32572269 |
| [11] | 潘志文, 高洁儿, 陈伟庭, 等. 基因组编辑技术在植物中的应用研究进展[J]. 贵州农业科学, 2020, 48(12): 16-20. |
| Pan ZW, Gao JE, Chen WT, et al. Research progress on application of genome editing technology in plants[J]. Guizhou Agric Sci, 2020, 48(12): 16-20. | |
| [12] |
袁伟曦, 喻云梅, 胡春财, 等. CRISPR/Cas9技术存在的问题及其改进措施的研究进展[J]. 生物技术通报, 2017, 33(4): 70-77.
doi: 10.13560/j.cnki.biotech.bull.1985.2017.04.009 |
| Yuan WX, Yu YM, Hu CC, et al. Current issues and progress in the application of CRISPR/Cas9 technique[J]. Biotechnol Bull, 2017, 33(4): 70-77. | |
| [13] |
Garcia JF, Parker R. MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system[J]. RNA, 2015, 21(8): 1393-1395.
doi: 10.1261/rna.051797.115 pmid: 26092944 |
| [14] |
Heinrich S, Sidler CL, Azzalin CM, et al. Stem-loop RNA labeling can affect nuclear and cytoplasmic mRNA processing[J]. RNA, 2017, 23(2): 134-141.
doi: 10.1261/rna.057786.116 pmid: 28096443 |
| [15] |
Heckl D, Charpentier E. Toward whole-transcriptome editing with CRISPR-Cas9[J]. Mol Cell, 2015, 58(4): 560-562.
doi: 10.1016/j.molcel.2015.05.016 pmid: 26000839 |
| [16] |
Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA[J]. EMBO J, 2012, 31(12): 2824-2832.
doi: 10.1038/emboj.2012.107 pmid: 22522703 |
| [17] |
Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease[J]. RNA, 2012, 18(4): 661-672.
doi: 10.1261/rna.030882.111 pmid: 22345129 |
| [18] |
Feng ZY, Mao YF, Xu NF, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis[J]. Proc Natl Acad Sci USA, 2014, 111(12): 4632-4637.
doi: 10.1073/pnas.1400822111 URL |
| [19] |
Zhong ZH, Sretenovic S, Ren QR, et al. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG[J]. Mol Plant, 2019, 12(7): 1027-1036.
doi: S1674-2052(19)30124-8 pmid: 30928637 |
| [20] | 雷建峰. 棉花高效CRISPR/Cas9基因编辑体系的研究[D]. 乌鲁木齐: 新疆农业大学, 2021. |
| Lei JF. Studies on high-efficient CRISPR/Cas9 gene editing system in cotton[D]. Urumqi: Xinjiang Agricultural University, 2021. | |
| [21] |
Erijman A, Dantes A, Bernheim R, et al. Transfer-PCR(TPCR): a highway for DNA cloning and protein engineering[J]. J Struct Biol, 2011, 175(2): 171-177.
doi: 10.1016/j.jsb.2011.04.005 pmid: 21515384 |
| [22] |
Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana[J]. Plant J, 1998, 16(6): 735-743.
doi: 10.1046/j.1365-313x.1998.00343.x pmid: 10069079 |
| [23] |
Gao W, Long L, Tian XQ, et al. Genome editing in cotton with the CRISPR/Cas9 system[J]. Front Plant Sci, 2017, 8: 1364.
doi: 10.3389/fpls.2017.01364 pmid: 28824692 |
| [24] |
Zeng DC, Li XT, Huang JF, et al. Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice[J]. Plant Biotechnol J, 2020, 18(6): 1348-1350.
doi: 10.1111/pbi.13293 pmid: 31696609 |
| [25] |
Li JY, Luo JM, Xu ML, et al. Plant genome editing using xCas9 with expanded PAM compatibility[J]. J Genet Genomics, 2019, 46(5): 277-280.
doi: S1673-8527(19)30052-9 pmid: 31054950 |
| [26] |
Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification[J]. Nat Biotechnol, 2014, 32(6): 577-582.
doi: 10.1038/nbt.2909 pmid: 24770324 |
| [27] |
Tsai SQ, Wyvekens N, Khayter C, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing[J]. Nat Biotechnol, 2014, 32(6): 569-576.
doi: 10.1038/nbt.2908 pmid: 24770325 |
| [28] |
Oh Y, Kim H, Kim SG. Virus-induced plant genome editing[J]. Curr Opin Plant Biol, 2021, 60: 101992.
doi: 10.1016/j.pbi.2020.101992 URL |
| [29] |
Idris AM, Brown JK. Cotton leaf crumple virus is a distinct western hemisphere begomovirus species with complex evolutionary relationships indicative of recombination and reassortment[J]. Phytopathology, 2004, 94(10): 1068-1074.
doi: 10.1094/PHYTO.2004.94.10.1068 pmid: 18943795 |
| [30] |
Gu ZH, Huang CJ, Li FF, et al. A versatile system for functional analysis of genes and microRNAs in cotton[J]. Plant Biotechnol J, 2014, 12(5): 638-649.
doi: 10.1111/pbi.12169 pmid: 24521483 |
| [31] |
Lei JF, Dai PH, Li Y, et al. Heritable gene editing using FT mobile guide RNAs and DNA viruses[J]. Plant Methods, 2021, 17(1): 20.
doi: 10.1186/s13007-021-00719-4 pmid: 33596981 |
| [32] |
赵燚, 雷建峰, 刘敏, 等. CLCrV介导的VIGE体系承载力的研究[J]. 生物技术通报, 2022, 38(11): 210-219.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0173 |
|
Zhao Y, Lei JF, Liu M, et al. Research on the carrying capacity of CLCrV-mediated VIGE system[J]. Biotechnol Bull, 2022, 38(11): 210-219.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0173 |
|
| [33] |
Ma XN, Zhang XY, Liu HM, et al. Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9[J]. Nat Plants, 2020, 6(7): 773-779.
doi: 10.1038/s41477-020-0704-5 pmid: 32601419 |
| [34] |
Ariga H, Toki S, Ishibashi K. Potato virus X vector-mediated DNA-free genome editing in plants[J]. Plant Cell Physiol, 2020, 61(11): 1946-1953.
doi: 10.1093/pcp/pcaa123 pmid: 32991731 |
| [35] |
Liu Q, Zhao CL, Sun K, et al. Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes[J]. Mol Plant, 2023, 16(3): 616-631.
doi: 10.1016/j.molp.2023.02.003 pmid: 36751129 |
| [36] |
Liu Y, Yang G, Huang SH, et al. Enhancing prime editing by Csy4-mediated processing of pegRNA[J]. Cell Res, 2021, 31(10): 1134-1136.
doi: 10.1038/s41422-021-00520-x pmid: 34103663 |
| [37] |
Jiang YY, Chai YP, Lu MH, et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize[J]. Genome Biol, 2020, 21(1): 257.
doi: 10.1186/s13059-020-02170-5 |
| [1] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [2] | 赵燚, 雷建峰, 刘敏, 胡子曜, 代培红, 刘超, 李月, 刘晓东. CLCrV介导的VIGE体系承载力的研究[J]. 生物技术通报, 2022, 38(11): 210-219. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||