








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 172-180.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0657
收稿日期:2024-07-10
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
武志强,男,博士,研究员,研究方向:植物细胞器基因组;E-mail: wuzhiqiang@caas.cn作者简介:周家伟,女,博士,研究方向:植物基因组;E-mail: jiaweizhou@webmail.hzau.edu.cn
基金资助:
ZHOU Jia-wei( ), WU Zhi-qiang(
), WU Zhi-qiang( )
)
Received:2024-07-10
Published:2024-10-26
Online:2024-11-20
摘要:
【目的】mitoTALENs植物线粒体基因编辑技术能够高效地实现线粒体基因的敲除,进而有效实现线粒体基因功能的研究,但mitoTALENs载体构建过程非常的繁琐复杂且目前仍没有较为系统完整的载体构建方法作为参考。为解决这个问题,结合前人已发表的及本实验室摸索出的方法对mitoTALENs载体构建的完整过程进行了详尽描述,为之后利用mitoTALENs技术进行植物线粒体基因功能研究的研究者们提供重要参考。【方法】以水稻线粒体WA352基因作为目的基因,利用其序列特异性区域设计了靶点TAL,首先采用Platinum gate TALEN assembly的两步组装技术分别构建了mitoTALENs的TALEN-left和TALEN-right载体,然后利用multisite LR反应将TALEN-left、TALEN-right及含有其他功能元件的进入载体和目的载体进行反应,生成最终的表达载体。【结果】第一步组装构建10个载体,第二步组装构建2个载体,最后通过multisite LR反应构建1个终表达载体。【结论】详细介绍了mitoTALENs载体的构建过程,为该技术使用者提供重要参考,以促进植物线粒体基因编辑研究领域的发展。
周家伟, 武志强. mitoTALENs植物线粒体基因编辑载体的构建方法[J]. 生物技术通报, 2024, 40(10): 172-180.
ZHOU Jia-wei, WU Zhi-qiang. Construction Method of mitoTALENs Mitochondrial Gene Editing Vector in Plants[J]. Biotechnology Bulletin, 2024, 40(10): 172-180.
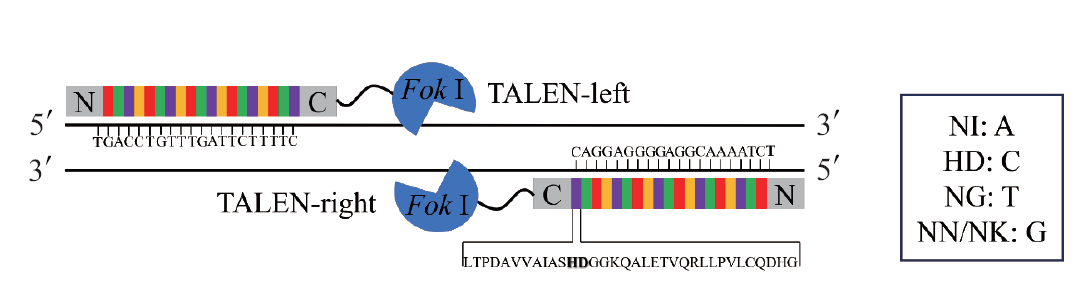
图1 TALENs的结构 加粗的HD及方框中的NI、HD、NG、NN和NK均表示重复可变双残基(RVD)
Fig. 1 Structure of TALENs The bold HD and the NI, HD, NG, NN, and NK in the box all indicate repeat variable diresidues(RVD)
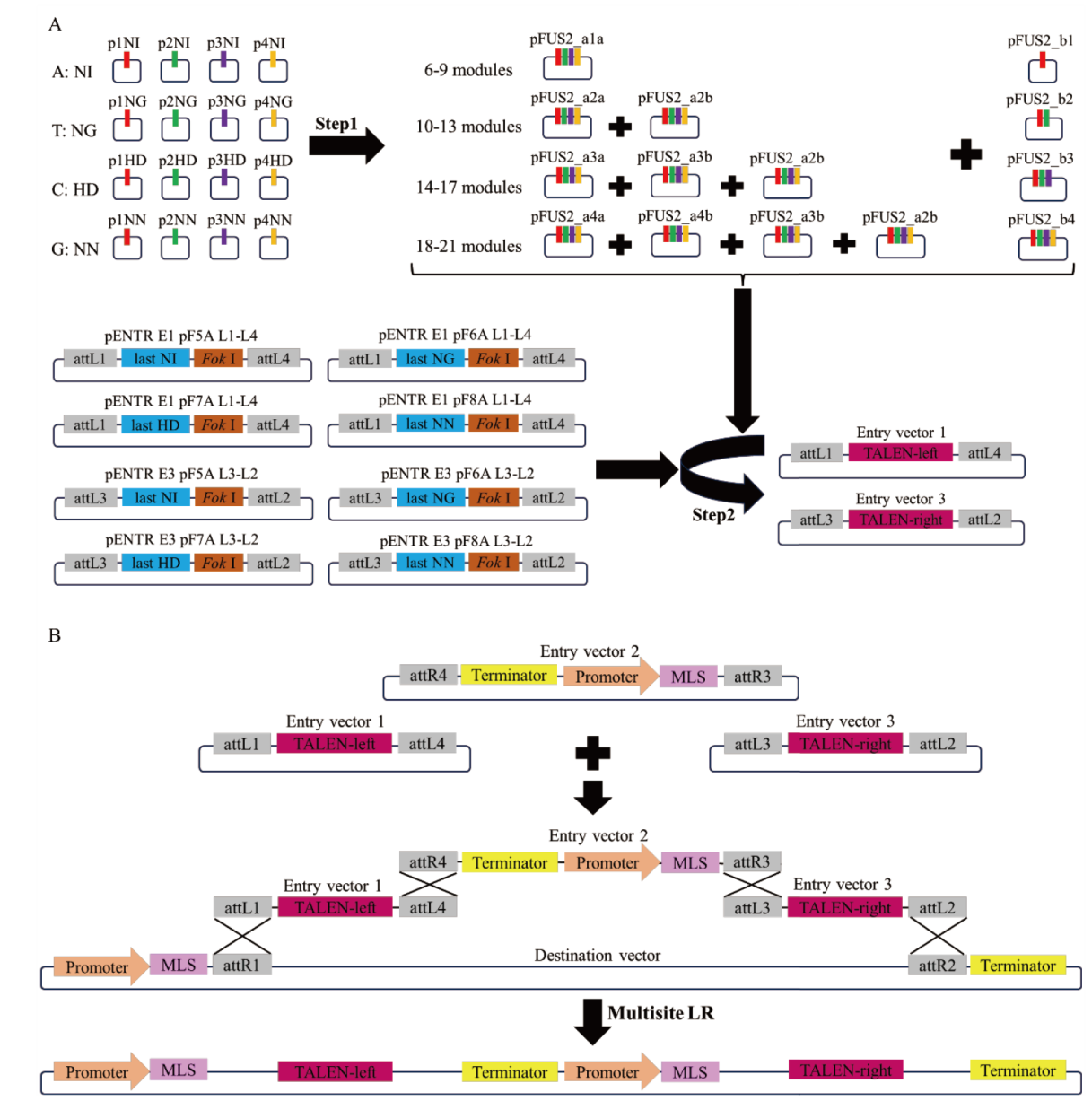
图3 mitoTALENs载体构建原理图 A:TALE阵列的两步组装原理图;B:multisite LR反应原理图
Fig. 3 Schematic diagram of mitoTALENs vector construction A: Two-step assembly schematic diagram of TALE array. B: Schematic diagram multisite LR reaction
| 载体类型 Vector type | 载体内容 Vector content | 载体原核抗性 Vector prokaryotic resistance |
|---|---|---|
| 模块质粒Module plasmids | p1NI,p2NI,p3NI,p4NI,p1NG,p2NG,p3NG,p4NG,p1HD,p2HD,p3HD,p4HD,p1NN,p2NN,p3NN,p4NN | AmpR |
| 中间阵列质粒 Intermediate array plasmids | pFUS2_a1a,pFUS2_a2a,pFUS2_a2b,pFUS2_a3a,pFUS2_a3b,pFUS2_a4a,pFUS2_a4b,pFUS2_b1,pFUS2_b2,pFUS2_b3,pFUS2_b4 | SpeR |
| 进入载体1 Entry vectors 1 | pENTR E1 pF5A L1-L4,pENTR E1 pF6A L1-L4, pENTRE1 pF7A L1-L4,pENTR E1 pF8A L1-L4 | KanR |
| 进入载体2 Entry vectors 2 | pENTER E2 HSPter-35Sp-mito | KanR |
| 进入载体3 Entry vectors 3 | pENTR E3 pR5A L3-L2,pENTR E3 pR6A L3-L2, pENTR E3 pR7A L3-L2,pENTR E3 pR8A L3-L2 | KanR |
| 目的载体 Destination vector | pDESTpK7WG2mito | SpeR |
表1 mitoTALENs载体构建所需的质粒载体
Table 1 Plasmid vectors required for the construction of mitoTALENs vector
| 载体类型 Vector type | 载体内容 Vector content | 载体原核抗性 Vector prokaryotic resistance |
|---|---|---|
| 模块质粒Module plasmids | p1NI,p2NI,p3NI,p4NI,p1NG,p2NG,p3NG,p4NG,p1HD,p2HD,p3HD,p4HD,p1NN,p2NN,p3NN,p4NN | AmpR |
| 中间阵列质粒 Intermediate array plasmids | pFUS2_a1a,pFUS2_a2a,pFUS2_a2b,pFUS2_a3a,pFUS2_a3b,pFUS2_a4a,pFUS2_a4b,pFUS2_b1,pFUS2_b2,pFUS2_b3,pFUS2_b4 | SpeR |
| 进入载体1 Entry vectors 1 | pENTR E1 pF5A L1-L4,pENTR E1 pF6A L1-L4, pENTRE1 pF7A L1-L4,pENTR E1 pF8A L1-L4 | KanR |
| 进入载体2 Entry vectors 2 | pENTER E2 HSPter-35Sp-mito | KanR |
| 进入载体3 Entry vectors 3 | pENTR E3 pR5A L3-L2,pENTR E3 pR6A L3-L2, pENTR E3 pR7A L3-L2,pENTR E3 pR8A L3-L2 | KanR |
| 目的载体 Destination vector | pDESTpK7WG2mito | SpeR |
| 模块质粒的个数 Number of module plasmids | 中间阵列质粒 Intermediate array plasmids | 模块质粒 Module plasmids | T4 DNA ligase reaction buffer | Bsa I-HF | Quick ligase | ddH2O | 总体积 Total volume/µL |
|---|---|---|---|---|---|---|---|
| 1 | 0.3 | 0.3×1 | 0.2 | 0.1 | 0.1 | 1 | 2 |
| 2 | 0.3 | 0.3×2 | 0.2 | 0.1 | 0.1 | 0.7 | 2 |
| 3 | 0.3 | 0.3×3 | 0.2 | 0.1 | 0.1 | 0.4 | 2 |
| 4 | 0.3 | 0.3×4 | 0.2 | 0.1 | 0.1 | 0.1 | 2 |
表2 第一步组装的反应体系
Table 2 Reaction system in the first step assembly
| 模块质粒的个数 Number of module plasmids | 中间阵列质粒 Intermediate array plasmids | 模块质粒 Module plasmids | T4 DNA ligase reaction buffer | Bsa I-HF | Quick ligase | ddH2O | 总体积 Total volume/µL |
|---|---|---|---|---|---|---|---|
| 1 | 0.3 | 0.3×1 | 0.2 | 0.1 | 0.1 | 1 | 2 |
| 2 | 0.3 | 0.3×2 | 0.2 | 0.1 | 0.1 | 0.7 | 2 |
| 3 | 0.3 | 0.3×3 | 0.2 | 0.1 | 0.1 | 0.4 | 2 |
| 4 | 0.3 | 0.3×4 | 0.2 | 0.1 | 0.1 | 0.1 | 2 |

图4 第一步组装方法示意图 A:WA352基因的靶点;B:TALE阵列的第一步组装方法
Fig. 4 Schematic diagram of the first step assembly method A: The target of WA352 gene. B: The first step assembly method for TALE arrays
| 模块质粒的个数 Number of module plasmids | pFUS2_a质粒 pFUS2_a plasmids | pFUS2_b质粒 pFUS2_b plasmids | 入门载体 Entry vectors | T4 DNA ligase reaction buffer | Esp3 I | Quick ligase | ddH2O | 总体积 Total volume/µL |
|---|---|---|---|---|---|---|---|---|
| 6-9 | 0.6×1 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 1.7 | 4 |
| 10-13 | 0.6×2 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 1.1 | 4 |
| 14-17 | 0.6×3 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 0.5 | 4 |
| 18-21 | 0.6×4 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 0 | 4.1 |
表3 第二步组装的反应体系
Table 3 Reaction system in the second step assembly
| 模块质粒的个数 Number of module plasmids | pFUS2_a质粒 pFUS2_a plasmids | pFUS2_b质粒 pFUS2_b plasmids | 入门载体 Entry vectors | T4 DNA ligase reaction buffer | Esp3 I | Quick ligase | ddH2O | 总体积 Total volume/µL |
|---|---|---|---|---|---|---|---|---|
| 6-9 | 0.6×1 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 1.7 | 4 |
| 10-13 | 0.6×2 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 1.1 | 4 |
| 14-17 | 0.6×3 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 0.5 | 4 |
| 18-21 | 0.6×4 | 0.6 | 0.3 | 0.4 | 0.2 | 0.2 | 0 | 4.1 |
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 目的Purpose |
|---|---|---|
| Step1_F | TTGATGCCTGGCAGTTCCCT | 第一步组装结果检测Step 1 assembly result detection |
| Step1_R | CGAACCGAACAGGCTTATGT | |
| Step2_F | GGACCGTCGCTGTCAAGTATCA | 第二步组装结果检测 Step 2 assembly result detection |
| Step2_R | AAGAACTCCATCACCTTCATCTCCAG | |
| attB1_F | ACAAGTTTGTACAAAAAAGCAGGCT | 终载体组装结果检测 Result detection of final vectorr assembly |
| attB4_R | CAACTTTGTATAGAAAAGTTGGGTGTCT | |
| attB4_F | AGACACCCAACTTTTCTATACAAAGTTG | |
| attB3_R | CAACTTTATTATACAAAGTTGTGGAATCGAGC | |
| attB3_F | GCTCGATTCCACAACTTTGTATAATAAAGTTG | |
| attB2_R | ACCACTTTGTACAAGAAAGCTGG |
表4 引物列表
Table 4 Primers list
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 目的Purpose |
|---|---|---|
| Step1_F | TTGATGCCTGGCAGTTCCCT | 第一步组装结果检测Step 1 assembly result detection |
| Step1_R | CGAACCGAACAGGCTTATGT | |
| Step2_F | GGACCGTCGCTGTCAAGTATCA | 第二步组装结果检测 Step 2 assembly result detection |
| Step2_R | AAGAACTCCATCACCTTCATCTCCAG | |
| attB1_F | ACAAGTTTGTACAAAAAAGCAGGCT | 终载体组装结果检测 Result detection of final vectorr assembly |
| attB4_R | CAACTTTGTATAGAAAAGTTGGGTGTCT | |
| attB4_F | AGACACCCAACTTTTCTATACAAAGTTG | |
| attB3_R | CAACTTTATTATACAAAGTTGTGGAATCGAGC | |
| attB3_F | GCTCGATTCCACAACTTTGTATAATAAAGTTG | |
| attB2_R | ACCACTTTGTACAAGAAAGCTGG |
| 目的基因 Target gene | 靶点 Target | 第一步组装 Step 1 | 第二步组装 Step 2 | 多位点LR Multisite LR |
|---|---|---|---|---|
| WA352 | TAL | pFUS2_a4a(L1), pFUS2_a4b(L2), pFUS2_a3b(L3), pFUS2_a2b(L4), pFUS2_b3(L5) | pENTR E1 pF5A L1-L4(last HD) | WA352::mitoTALEN |
| pFUS2_a4a(R1), pFUS2_a4b(R2), pFUS2_a3b(R3), pFUS2_a2b(R4), pFUS2_b3(R5) | pENTR E3 pR5A L3-L2(last HD) | |||
| 总数Total | 10 | 2 | 1 |
表5 本研究构建的载体
Table 5 Vectors constructed in this study
| 目的基因 Target gene | 靶点 Target | 第一步组装 Step 1 | 第二步组装 Step 2 | 多位点LR Multisite LR |
|---|---|---|---|---|
| WA352 | TAL | pFUS2_a4a(L1), pFUS2_a4b(L2), pFUS2_a3b(L3), pFUS2_a2b(L4), pFUS2_b3(L5) | pENTR E1 pF5A L1-L4(last HD) | WA352::mitoTALEN |
| pFUS2_a4a(R1), pFUS2_a4b(R2), pFUS2_a3b(R3), pFUS2_a2b(R4), pFUS2_b3(R5) | pENTR E3 pR5A L3-L2(last HD) | |||
| 总数Total | 10 | 2 | 1 |
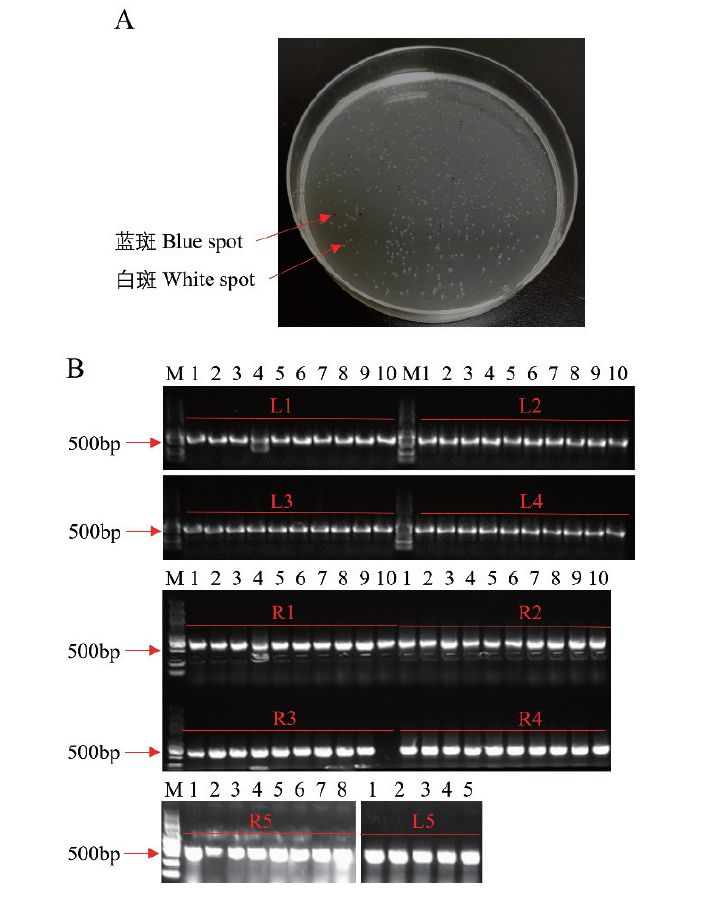
图5 第一步组装结果的阳性检测 A:蓝白斑筛选阳性单克隆;B:L1-L5和R1-R5这10个载体的PCR阳性检测
Fig. 5 Positive detection of assembly results in the first step A: Screening positive monoclonal via blue and white spot. B: The ten vectors L1-L5 and R1-R5 were detected by PCR
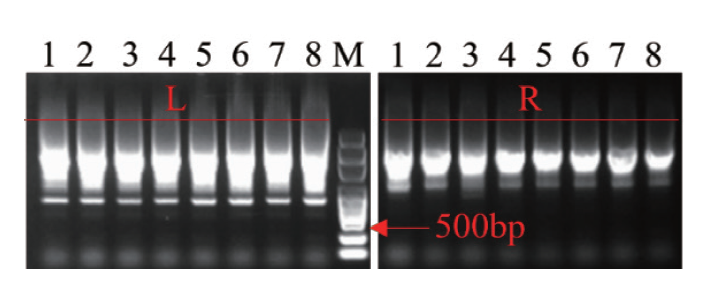
图6 第二步组装结果的PCR检测 WA352::TALEN-left(L)和WA352::TALEN-right(R)的PCR检测
Fig. 6 PCR detection of assembly results in the second step PCR detection of WA352::TALEN-left(L)and WA352::TALEN-right(R)

图7 第二步组装结果的酶切检测 A:WA352基因的TAL-left(L)和TAL-right(R)所对应的重复可变双残基(RVDs),其中NI是Msc I的消化位点;B:TAL-left(L)和TAL-right(R)经Msc I消化后的产物胶图
Fig. 7 Enzyme digestion of the assembly results in the second step A: The repeat variable diresidues(RVDs)corresponding to TAL-left(L)and TAL-right(R)of WA352 gene, where NI is the digestion site of Msc I. B: Gel image of the products of TAL-left(L)and TAL-right(R)digested by Msc I
| [1] |
Gao CX. Genome engineering for crop improvement and future agriculture[J]. Cell, 2021, 184(6): 1621-1635.
doi: 10.1016/j.cell.2021.01.005 pmid: 33581057 |
| [2] | Chang YZ, Liu BL, Jiang YY, et al. Induce male sterility by CRISPR/Cas9-mediated mitochondrial genome editing in tobacco[J]. Funct Integr Genomics, 2023, 23(3): 205. |
| [3] |
Kazama T, Okuno M, Watari Y, et al. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing[J]. Nat Plants, 2019, 5(7): 722-730.
doi: 10.1038/s41477-019-0459-z pmid: 31285556 |
| [4] | Arimura SI, Ayabe H, Sugaya H, et al. Targeted gene disruption of ATP synthases 6-1 and 6-2 in the mitochondrial genome of Arabidopsis thaliana by mitoTALENs[J]. Plant J, 2020, 104(6): 1459-1471. |
| [5] |
Omukai S, Arimura SI, Toriyama K, et al. Disruption of mitochondrial open reading frame 352 partially restores pollen development in cytoplasmic male sterile rice[J]. Plant Physiol, 2021, 187(1): 236-246.
doi: 10.1093/plphys/kiab236 pmid: 34015134 |
| [6] | Takatsuka A, Kazama T, Arimura SI, et al. TALEN-mediated depletion of the mitochondrial gene orf312 proves that it is a Tadukan-type cytoplasmic male sterility-causative gene in rice[J]. Plant J, 2022, 110(4): 994-1004. |
| [7] |
Kuwabara K, Arimura SI, Shirasawa K, et al. orf137 triggers cytoplasmic male sterility in tomato[J]. Plant Physiol, 2022, 189(2): 465-468.
doi: 10.1093/plphys/kiac082 pmid: 35212743 |
| [8] |
Forner J, Kleinschmidt D, Meyer EH, et al. Targeted knockout of a conserved plant mitochondrial gene by genome editing[J]. Nat Plants, 2023, 9(11): 1818-1831.
doi: 10.1038/s41477-023-01538-2 pmid: 37814021 |
| [9] | Ayabe H, Toyoda A, Iwamoto A, et al. Mitochondrial gene defects in Arabidopsis can broadly affect mitochondrial gene expression through copy number[J]. Plant Physiol, 2023, 191(4): 2256-2275. |
| [10] | Xu FY, Su TB, Zhang XC, et al. Editing of ORF138 restores fertility of Ogura cytoplasmic male sterile broccoli via mitoTALENs[J]. Plant Biotechnol J, 2024, 22(5): 1325-1334. |
| [11] |
Nicolia A, Scotti N, D'Agostino N, et al. Mitochondrial DNA editing in potato through mitoTALEN and mitoTALECD: molecular characterization and stability of editing events[J]. Plant Methods, 2024, 20(1): 4.
doi: 10.1186/s13007-023-01124-9 pmid: 38183104 |
| [12] | Zhou JW, Nie LY, Zhang S, et al. Mitochondrial genome editing of WA352 via mitoTALENs restore fertility in cytoplasmic male sterile rice[J]. Plant Biotechnol J, 2024, 22(7): 1960-1962. |
| [13] |
Gaj T, Gersbach CA, Barbas CF III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering[J]. Trends Biotechnol, 2013, 31(7): 397-405.
doi: 10.1016/j.tibtech.2013.04.004 pmid: 23664777 |
| [14] |
Khalil AM. The genome editing revolution: review[J]. J Genet Eng Biotechnol, 2020, 18(1): 68.
doi: 10.1186/s43141-020-00078-y pmid: 33123803 |
| [15] | Arimura SI. MitoTALENs: a method for targeted gene disruption in plant mitochondrial genomes[J]. Methods Mol Biol, 2022, 2363: 335-340. |
| [16] |
Sakuma T, Ochiai H, Kaneko T, et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity[J]. Sci Rep, 2013, 3: 3379.
doi: 10.1038/srep03379 pmid: 24287550 |
| [17] |
Sakuma T, Yamamoto T. Engineering customized TALENs using the platinum gate TALEN kit[J]. Methods Mol Biol, 2016, 1338: 61-70.
doi: 10.1007/978-1-4939-2932-0_6 pmid: 26443214 |
| [18] |
Guo JY, Zhang X, Chen XX, et al. Precision modeling of mitochondrial diseases in zebrafish via DdCBE-mediated mtDNA base editing[J]. Cell Discov, 2021, 7(1): 78.
doi: 10.1038/s41421-021-00307-9 pmid: 34480028 |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||