








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 156-167.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0307
王亚萍1( ), 金兰1, 郝金凤2, 长明1, 王艳丹1, 高峰1(
), 金兰1, 郝金凤2, 长明1, 王艳丹1, 高峰1( )
)
收稿日期:2025-03-21
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
高峰,男,博士,副教授,研究方向 :植物分子生物学;E-mail: imgaofeng@163.com作者简介:王亚萍,女,硕士研究生,研究方向 :生物化学与分子生物学;E-mail: imypwang@163.com
基金资助:
WANG Ya-ping1( ), JIN Lan1, HAO Jin-feng2, CHANG Ming1, WANG Yan-dan1, GAO Feng1(
), JIN Lan1, HAO Jin-feng2, CHANG Ming1, WANG Yan-dan1, GAO Feng1( )
)
Received:2025-03-21
Published:2025-12-26
Online:2026-01-06
摘要:
目的 RGLG蛋白属于RING(Really Interesting New Gene)型E3泛素连接酶,通过泛素化降解其他蛋白参与植物生长发育和非生物胁迫响应。鉴定甜瓜CmRGLG基因家族(CmRGLGs)成员,并分析相关基因的表达模式,为进一步探究其潜在功能奠定理论基础。 方法 采用生物信息学方法对CmRGLGs染色体定位、基因结构及其编码蛋白的理化特性、系统进化、共线性关系、蛋白互作等方面进行分析,通过RT-qPCR对各成员在不同器官、不同激素浓度梯度和非生物胁迫处理后甜瓜幼叶中的表达水平进行解析。 结果 在甜瓜全基因组中共鉴定出6个CmRGLGs成员,根据各基因在染色体上的位置及排列顺序依次命名为CmRGLG1-CmRGLG6;所编码的蛋白均为亲水性蛋白,氨基酸数为364-596 aa;在进行系统进化分析时,CmRGLGs基因分属于不同的3个分支。种内与种间共线性分析表明,CmRGLGs中不存在基因重复事件,CmRGLG1、CmRGLG2、CmRGLG4、CmRGLG5在拟南芥、黄瓜、番茄中均存在共线基因;启动子区中存在与植物激素和非生物胁迫响应有关的顺式作用元件;蛋白互作网络预测分析提示,CmRGLGs相互作用的蛋白主要富集在泛素‒蛋白转移酶活性、蛋白质代谢、生物合成和有机环状化合物结合等途径;表达特性分析结果显示,CmRGLG5和CmRGLG6在茎中的表达量最低,CmRGLG1、CmRGLG2、CmRGLG3、CmRGLG4在花中的表达量较高,且各基因成员在不同植物激素和非生物胁迫中具有不同程度的表达。 结论 甜瓜CmRGLGs在不同植物激素处理条件下,多数成员的表达水平有下调趋势;在非生物胁迫处理条件下,多数成员的表达水平呈先上调再下调的趋势。
王亚萍, 金兰, 郝金凤, 长明, 王艳丹, 高峰. 甜瓜CmRGLG基因家族鉴定及表达特性分析[J]. 生物技术通报, 2025, 41(12): 156-167.
WANG Ya-ping, JIN Lan, HAO Jin-feng, CHANG Ming, WANG Yan-dan, GAO Feng. Identification and Expression Characteristics Analysis of CmRGLG Gene Family in Melon[J]. Biotechnology Bulletin, 2025, 41(12): 156-167.
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Product size (bp) |
|---|---|---|---|
| CmGAPDH | ATCATTCCTAGCAGCACTGG | TTGGCATCAAATATGCTTGACCTG | 278 |
| CmRGLG1 | AGTGGGGTCATTATGGTTATCCG | CCTCTTTCTTGTCTCAGGGGCT | 116 |
| CmRGLG2 | GGTCAATCCCCCCCTTAT | GCTCCTGTTACCTCCTCCAA | 221 |
| CmRGLG3 | CTTGTTTCGGTTTTGGTGAT | CCAGGTGGTGTTTTAGGATTTC | 262 |
| CmRGLG4 | ATATCTCCCCAGAGAGTTGCA | AAGACCAGGTAGGCTTTGAAT | 97 |
| CmRGLG5 | CAAACTCCACAACAACCACGG | GAGATTAGATGACTCAAGTCCAGCG | 123 |
| CmRGLG6 | CGATTCTCCTAATCCCTACCA | TTGCCCACCACTCTTCTCTA | 280 |
表1 RT-qPCR引物序列
Table 1 RT-qPCR primer sequences
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Product size (bp) |
|---|---|---|---|
| CmGAPDH | ATCATTCCTAGCAGCACTGG | TTGGCATCAAATATGCTTGACCTG | 278 |
| CmRGLG1 | AGTGGGGTCATTATGGTTATCCG | CCTCTTTCTTGTCTCAGGGGCT | 116 |
| CmRGLG2 | GGTCAATCCCCCCCTTAT | GCTCCTGTTACCTCCTCCAA | 221 |
| CmRGLG3 | CTTGTTTCGGTTTTGGTGAT | CCAGGTGGTGTTTTAGGATTTC | 262 |
| CmRGLG4 | ATATCTCCCCAGAGAGTTGCA | AAGACCAGGTAGGCTTTGAAT | 97 |
| CmRGLG5 | CAAACTCCACAACAACCACGG | GAGATTAGATGACTCAAGTCCAGCG | 123 |
| CmRGLG6 | CGATTCTCCTAATCCCTACCA | TTGCCCACCACTCTTCTCTA | 280 |

图1 甜瓜CmRGLGs基因结构和保守基序分析A:基因结构;B:保守基序;C:基序序列徽标(每个字母的高度表示该位置氨基酸出现的频率)
Fig. 1 Gene structure and conservative motif analysis of CmRGLGs in melonA: Gene structure. B: Conserved motifs. C: Motif sequence logo (The height of each letter indicates the frequency of amino acid occurrence at that position)
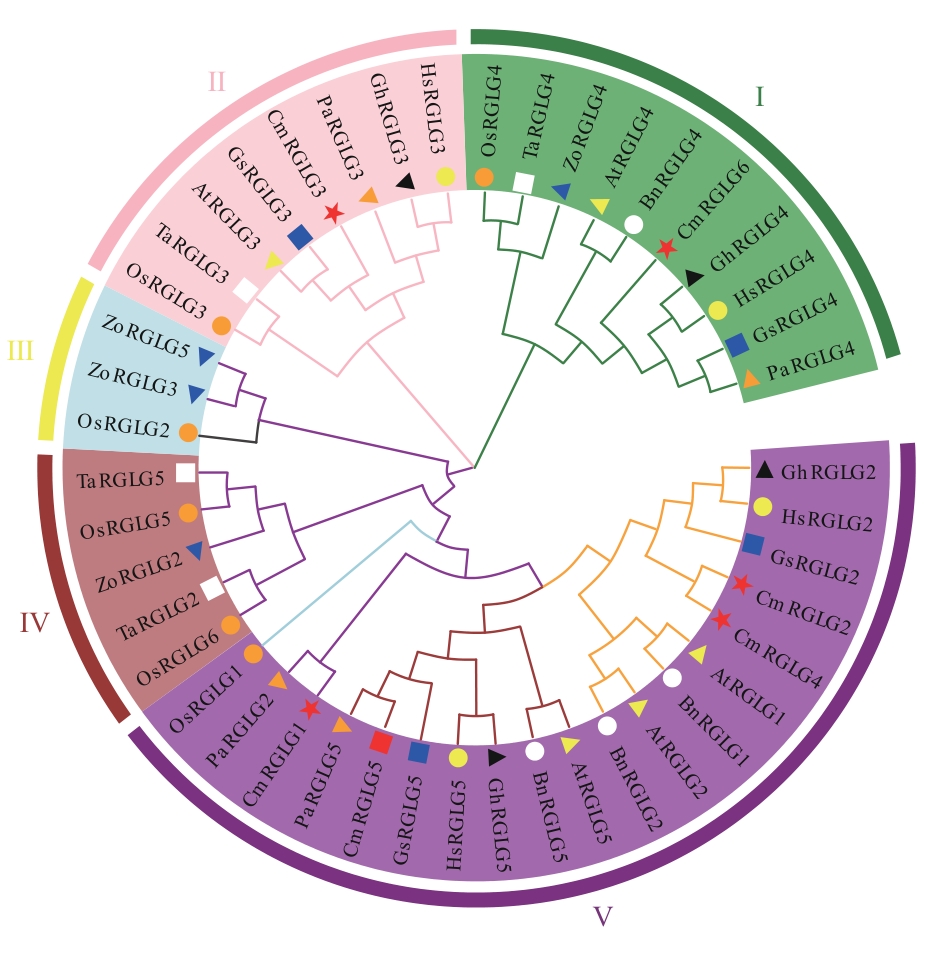
图2 甜瓜与其他物种RGLG蛋白家族系统进化树Cm:甜瓜;At:拟南芥;Os:水稻;Pa:银白杨;Ta:小麦;Gs:野大豆;Gh:陆地棉;Bn:甘蓝型油菜;Hs:木槿;Zo:姜
Fig. 2 Phylogenetic tree of RGLG protein family in melon and other speciesCm: Cucumis melo; At: Arabidopsis thaliana; Os: Oryza sativa; Pa: Populus alba; Ta: Triticum aestivum; Gs: Glycine soja; Gh: Gossypium hirsutum; Bn: Brassica napus; Hs: Hibiscus syriacus; Zo: Zingiber officinale
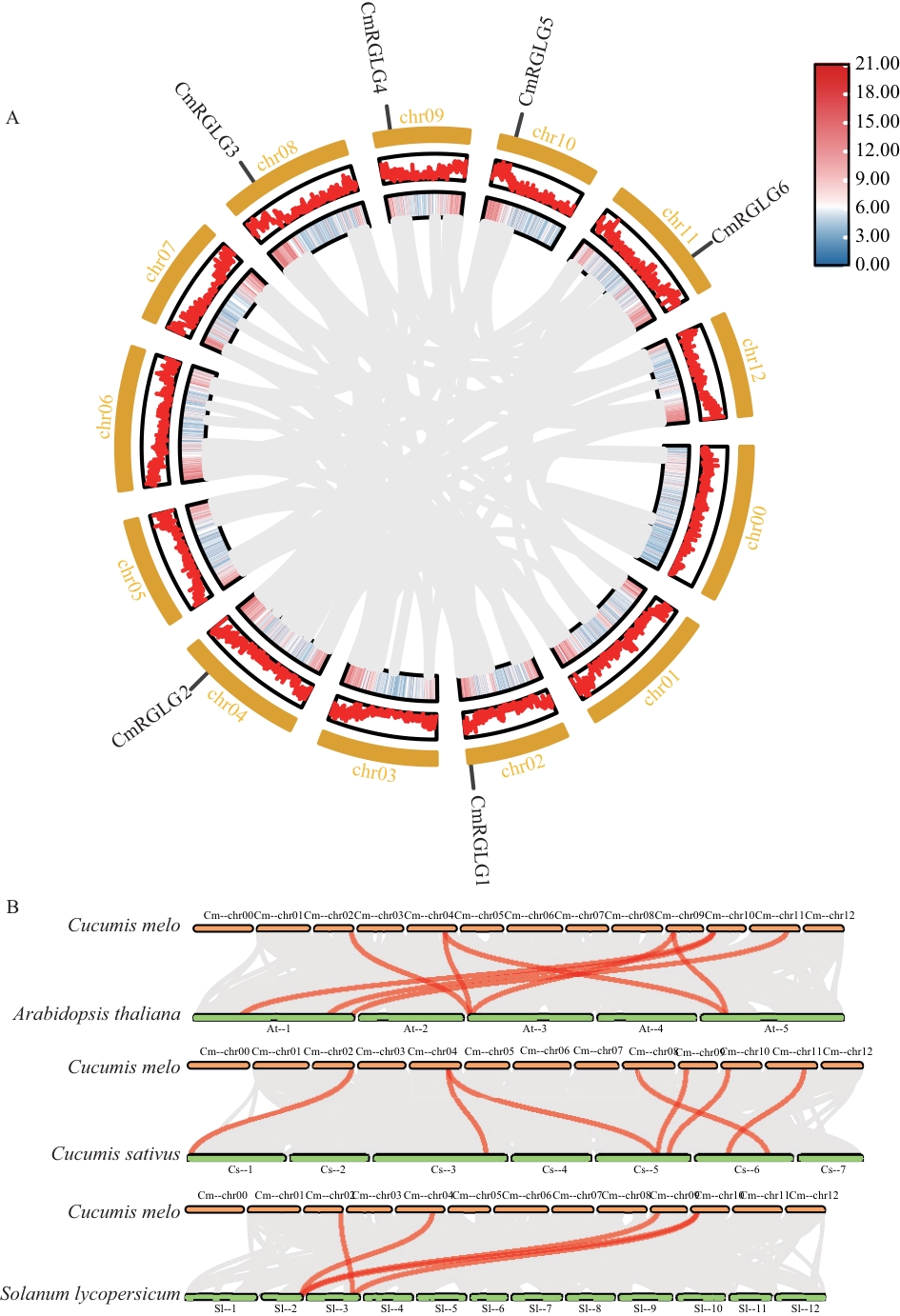
图4 甜瓜CmRGLGs种内(A)与不同物种间(B)共线性分析灰色线条代表甜瓜基因组与参与分析物种之间的共线性区块;红色线条突出了RGLG基因对
Fig. 4 Intraspecific (A) and interspecific synteny (B) analysis of CmRGLGs in melonGray lines indicate collinear blocks between the melon genome and the genomes of the analyzed species; red lines highlight RGLG gene pairs
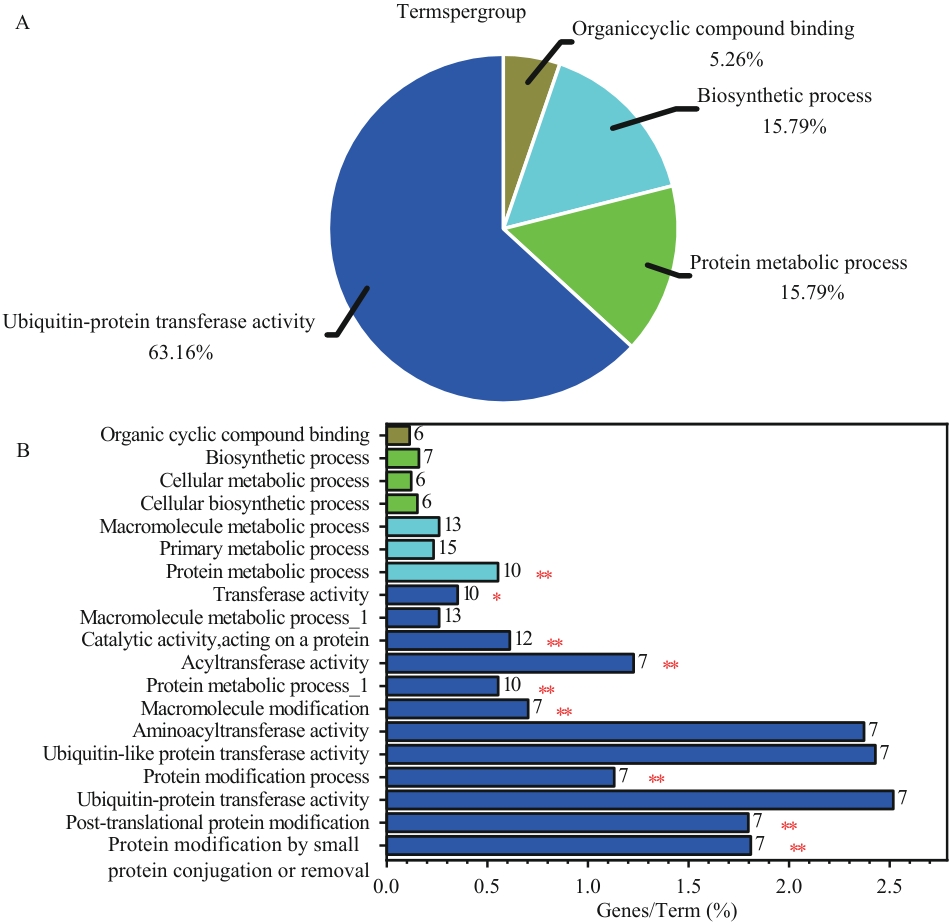
图6 Cytoscape 3.10.2分析蛋白互作中KEGG和GO注释统计结果A:KEGG通路富集分析;B:GO功能富集分析(横坐标表示每个Term中富集基因所占百分比,柱状图末端数字表示对应Term的具体基因数量;纵坐标表示显著富集的GO Terms)。*P<0.05,**P<0.01
Fig. 6 Statistical results of KEGG and GO annotation in protein-protein interaction analysis by Cytoscape 3.10.2A: KEGG pathway enrichment analysis. B: GO functional enrichment analysis (X-axis: percentage of enriched genes per Term; numbers at the end of bars indicate the specific gene count in each Term; Y-axis: significantly enriched GO Terms). *P<0.05, **P<0.01
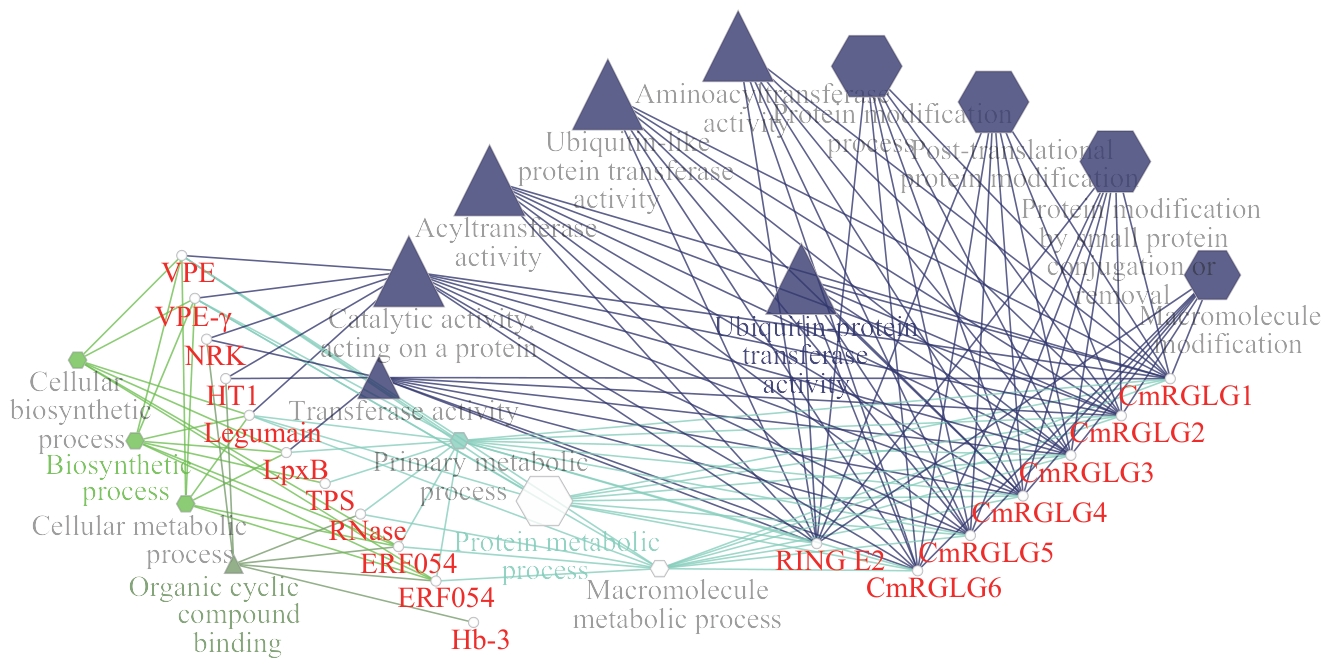
图7 甜瓜CmRGLGs蛋白相互作用网络六边形表示生物过程;三角形表示分子功能;圆形表示蛋白
Fig. 7 Protein-protein interaction network of CmRGLGs in melonHexagons indicate biological processes; triangles indicate molecular functions; circles indicate proteins
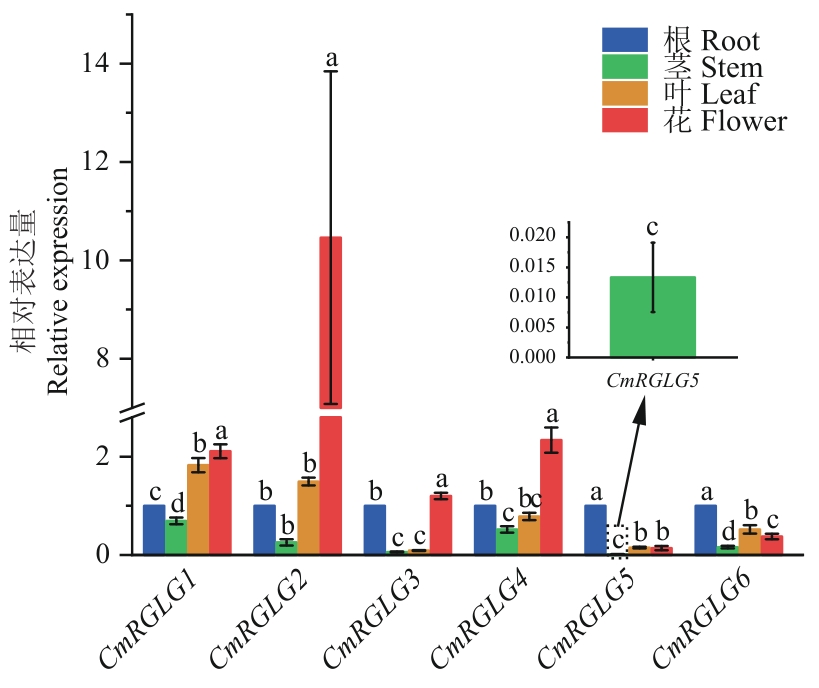
图8 甜瓜CmRGLGs在不同器官中的表达特性分析不同小写字母表示差异显著(P<0.05)。下同
Fig. 8 Analysis of expression characteristics of CmRGLGs in different organs of melonDifferent lowercase letters indicate significant differences (P<0.05). The same below

图9 甜瓜CmRGLGs在不同植物激素(A)和非生物胁迫处理(B)下的表达特性分析
Fig. 9 Analysis of expression characteristics of CmRGLGs under phytohormone (A) and abiotic stress treatments (B) in melon
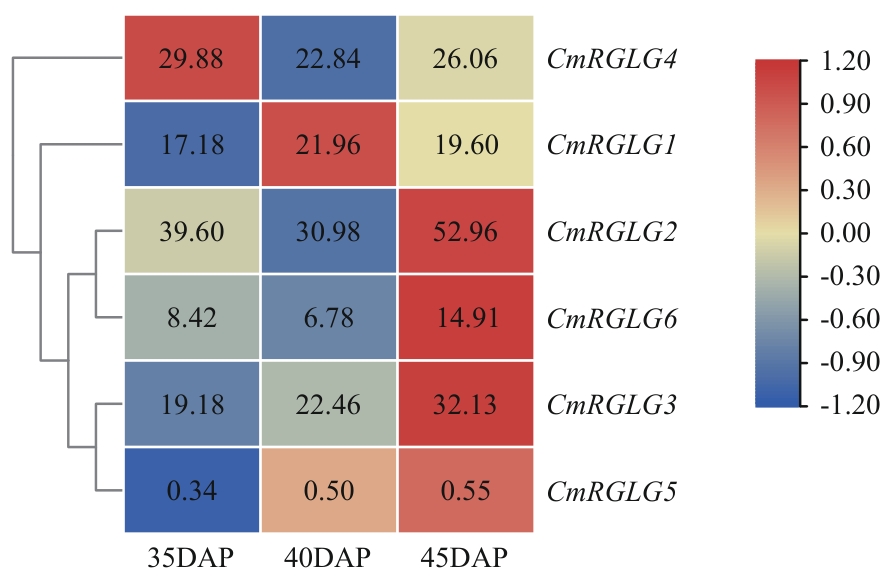
图10 甜瓜果实转录组数据中CmRGLGs的表达特性分析35、40、45 DAP分别表示授粉后35、40、45 d
Fig. 10 Analysis of expression characteristics of CmRGLGs in the transcriptome data of melon fruits35, 40, and 45 DAP indicate 35, 40, and 45 d after pollination (DAP), respectively
| [1] | Lobaina DP, Tarazi R, Castorino T, et al. The ubiquitin-proteasome system (UPS) and viral infection in plants [J]. Plants, 2022, 11(19): 2476. |
| [2] | Chen X, Dou QP, Liu JB, et al. Targeting ubiquitin-proteasome system with copper complexes for cancer therapy [J]. Front Mol Biosci, 2021, 8: 649151. |
| [3] | Yang Q, Zhao JY, Chen D, et al. E3 ubiquitin ligases: styles, structures and functions [J]. Mol Biomed, 2021, 2(1): 23. |
| [4] | Lee JH, Kim WT. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis [J]. Mol Cells, 2011, 31(3): 201-208. |
| [5] | Du Z, Zhou X, Li L, et al. plantsUPS: a database of plants’ ubiquitin proteasome system [J]. BMC Genomics, 2009, 10: 227. |
| [6] | Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases [J]. Annu Rev Biochem, 2009, 78: 399-434. |
| [7] | Saxena H, Negi H, Sharma B. Role of F-box E3-ubiquitin ligases in plant development and stress responses [J]. Plant Cell Rep, 2023, 42(7): 1133-1146. |
| [8] | Sharma B, Saxena H, Negi H. Genome-wide analysis of HECT E3 ubiquitin ligase gene family in Solanum lycopersicum [J]. Sci Rep, 2021, 11(1): 15891. |
| [9] | 苌兴超, 王雪松, 张沿政, 等. 大豆GmPUB32基因的克隆及耐盐功能分析 [J]. 中国油料作物学报, 2021, 43(4): 638-647. |
| Chang XC, Wang XS, Zhang YZ, et al. Cloning and salt resistance analysis of soybean GmPUB32 gene [J]. Chin J Oil Crop Sci, 2021, 43(4): 638-647. | |
| [10] | 徐焕焕, 张芳毓, 刘茜, 等. 番茄SlSL1重组蛋白的表达、纯化及体外泛素化活性检测 [J]. 合肥工业大学学报: 自然科学版, 2022, 45(8): 1130-1134. |
| Xu HH, Zhang FY, Liu Q, et al. Expression, purification and in vitro ubiquitination analysis of tomato SlSL1 recombinant protein [J]. J Hefei Univ Technol Nat Sci, 2022, 45(8): 1130-1134. | |
| [11] | Freemont PS. Ubiquitination: RING for destruction [J]. Curr Biol, 2000, 10(2): R84-R87. |
| [12] | Pan IC, Tsai HH, Cheng YT, et al. Post-transcriptional coordination of the Arabidopsis iron deficiency response is partially dependent on the E3 ligases RING domain ligase1 (RGLG1) and RING domain ligase2 (RGLG2) [J]. Mol Cell Proteom, 2015, 14(10): 2733-2752. |
| [13] | Stone SL, Hauksdóttir H, Troy A, et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis [J]. Plant Physiol, 2005, 137(1): 13-30. |
| [14] | Katoh S, Hong C, Tsunoda Y, et al. High precision NMR structure and function of the RING-H2 finger domain of EL5, a rice protein whose expression is increased upon exposure to pathogen-derived oligosaccharides [J]. J Biol Chem, 2003, 278(17): 15341-15348. |
| [15] | Li YZ, Wu BJ, Yu YL, et al. Genome-wide analysis of the RING finger gene family in apple [J]. Mol Genet Genomics, 2011, 286(1): 81-94. |
| [16] | Qi ZY, Ahammed GJ, Jiang CY, et al. The E3 ubiquitin ligase gene SlRING1 is essential for plant tolerance to cadmium stress in Solanum lycopersicum [J]. J Biotechnol, 2020, 324: 239-247. |
| [17] | Tan B, Lian XD, Cheng J, et al. Genome-wide identification and transcriptome profiling reveal that E3 ubiquitin ligase genes relevant to ethylene, auxin and abscisic acid are differentially expressed in the fruits of melting flesh and stony hard peach varieties [J]. BMC Genomics, 2019, 20(1): 892. |
| [18] | Tang XL, Mei YY, He KX, et al. The RING-type E3 ligase RIE1 sustains leaf longevity by specifically targeting AtACS7 to fine-tune ethylene production in Arabidopsis [J]. Proc Natl Acad Sci USA, 2024, 121(48): e2411271121. |
| [19] | Xin TX, Zhang Z, Li S, et al. Genetic regulation of ethylene dosage for cucumber fruit elongation [J]. Plant Cell, 2019, 31(5): 1063-1076. |
| [20] | Wu T, Wang Y, Jin J, et al. Soybean RING-type E3 ligase GmCHYR16 ubiquitinates the GmERF71 transcription factor for degradation to negatively regulate bicarbonate stress tolerance [J]. New Phytol, 2025, 246(3): 1128-1146. |
| [21] | Shahwar D, Khan Z, Park Y. Molecular marker-assisted mapping, candidate gene identification, and breeding in melon (Cucumis melo L.): a review [J]. Int J Mol Sci, 2023, 24(20): 15490. |
| [22] | Liu YW, Gao YY, Chen MX, et al. GIFTdb: a useful gene database for plant fruit traits improving [J]. Plant J, 2023, 116(4): 1030-1040. |
| [23] | Li ML, Dong XY, Long GZ, et al. Genome-wide analysis of Q-type C2H2 ZFP genes in response to biotic and abiotic stresses in sugar beet [J]. Biology, 2023, 12(10): 1309. |
| [24] | Nystrom SL, McKay DJ. Memes: A motif analysis environment in R using tools from the MEME Suite [J]. PLoS Comput Biol, 2021, 17(9): e1008991. |
| [25] | Zhao DB, Gao FJ, Guan PY, et al. Identification and analysis of differentially expressed trihelix genes in maize (Zea mays) under abiotic stresses [J]. PeerJ, 2023, 11: e15312. |
| [26] | 洪天澍, 海英, 恩和巴雅尔, 等. 甜瓜CmABCG8基因的表达特性分析 [J]. 生物技术通报, 2022, 38(7): 178-185. |
| Hong TS, Hai Y, En H, et al. Analysis of expression characteristics of CmABCG8 gene in Cucumis melo L [J]. Biotechnol Bull, 2022, 38(7): 178-185. | |
| [27] | Chen Q, Liu RJ, Wu YR, et al. ERAD-related E2 and E3 enzymes modulate the drought response by regulating the stability of PIP2 aquaporins [J]. Plant Cell, 2021, 33(8): 2883-2898. |
| [28] | Hao DD, Jin L, Wen X, et al. The RING E3 ligase SDIR1 destabilizes EBF1/EBF2 and modulates the ethylene response to ambient temperature fluctuations in Arabidopsis [J]. Proc Natl Acad Sci USA, 2021, 118(6): e2024592118. |
| [29] | Wang JY, Wang RY, Fang H, et al. Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice [J]. Mol Plant, 2021, 14(2): 253-266. |
| [30] | Wu Q, Zhang X, Peirats-Llobet M, et al. Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA [J]. Plant Cell, 2016, 28(9): 2178-2196. |
| [31] | 舒合凤, 胡荣美, 朱灵, 等. 水稻泛素连接酶基因RGLG31的同源克隆及表达分析[J/OL]. 分子植物育种, 2025. . |
| Shu HF, Hu RM, Zhu L, et al. Homologous cloning and expression analysis of rice ubiquitin ligase gene RGLG31[J/OL]. Mol Plant Breed, 2025. . | |
| [32] | Ma JF, Wang RB, Zhao HY, et al. Genome-wide characterization of the VQ genes in Triticeae and their functionalization driven by polyploidization and gene duplication events in wheat [J]. Int J Biol Macromol, 2023, 243: 125264. |
| [33] | Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure [J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192. |
| [34] | Sun WQ, Li MD, Wang JB. Genome-wide identification and characterization of the RCI2 gene family in allotetraploid Brassica napus compared with its diploid progenitors [J]. Int J Mol Sci, 2022, 23(2): 614. |
| [35] | Nagels Durand A, Iñigo S, Ritter A, et al. The Arabidopsis iron-sulfur protein GRXS17 is a target of the ubiquitin E3 ligases RGLG3 and RGLG4 [J]. Plant Cell Physiol, 2016, 57(9): 1801-1813. |
| [36] | Zhang X, Wu Q, An CC. RGLG3 and RGLG4, novel ubiquitin ligases modulating jasmonate signaling [J]. Plant Signal Behav, 2012, 7(12): 1709-1711. |
| [37] | Zhang X, Wu Q, Ren J, et al. Two novel RING-type ubiquitin ligases, RGLG3 and RGLG4, are essential for jasmonate-mediated responses in Arabidopsis [J]. Plant Physiol, 2012, 160(2): 808-822. |
| [38] | 陈佳琳. E3泛素连接酶CsRGLG4与转录因子CsAP2L调控柑橘果实采后绿霉病抗病性的机制研究 [D]. 重庆: 西南大学, 2024. |
| Chen JL. Mechanism of E3 ubiquitin ligase CsRGLG4 and transcription factor CsAP2L in regulating postharvest green mold resistance in citrus fruits [D]. Chongqing: Southwest University, 2024. | |
| [39] | Cheng MC, Hsieh EJ, Chen JH, et al. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response [J]. Plant Physiol, 2012, 158(1): 363-375. |
| [40] | Dong CH, Agarwal M, Zhang Y, et al. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1 [J]. Proc Natl Acad Sci USA, 2006, 103(21): 8281-8286. |
| [1] | 王芳, 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍, 姜亮. 植物和细菌TurboID邻近蛋白标记方法的建立[J]. 生物技术通报, 2025, 41(9): 44-53. |
| [2] | 陈强, 于璎霏, 张颖, 张冲. 茉莉酸甲酯对薄皮甜瓜‘绿宝石’采后冷害的调控[J]. 生物技术通报, 2025, 41(9): 105-114. |
| [3] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [4] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [5] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [6] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [7] | 黄国栋, 邓宇星, 程宏伟, 但焱南, 周会汶, 吴兰花. 大豆ZIP基因家族鉴定及响应铝胁迫的表达分析[J]. 生物技术通报, 2025, 41(9): 71-81. |
| [8] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [9] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [10] | 化文平, 刘菲, 浩佳欣, 陈尘. 丹参ADH基因家族的鉴定与表达模式分析[J]. 生物技术通报, 2025, 41(8): 211-219. |
| [11] | 腊贵晓, 赵玉龙, 代丹丹, 余永亮, 郭红霞, 史贵霞, 贾慧, 杨铁钢. 红花质膜H+-ATPase基因家族成员鉴定及响应低氮低磷胁迫的表达分析[J]. 生物技术通报, 2025, 41(8): 220-233. |
| [12] | 程雪, 付颖, 柴晓娇, 王红艳, 邓欣. 谷子LHC基因家族鉴定及非生物胁迫表达分析[J]. 生物技术通报, 2025, 41(8): 102-114. |
| [13] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [14] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [15] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||