








生物技术通报 ›› 2021, Vol. 37 ›› Issue (10): 186-195.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0197
仲建锋1( ), 李兴奎2, 徐重新1, 张霄1, 刘贤金1(
), 李兴奎2, 徐重新1, 张霄1, 刘贤金1( )
)
收稿日期:2021-02-20
出版日期:2021-10-26
发布日期:2021-11-12
作者简介:仲建锋,男,博士,副研究员,研究方向:农产品质量安全与生物控制技术;E-mail: 基金资助:
ZHONG Jian-feng1( ), LI Xing-kui2, XU Chong-xin1, ZHANG Xiao1, LIU Xian-jin1(
), LI Xing-kui2, XU Chong-xin1, ZHANG Xiao1, LIU Xian-jin1( )
)
Received:2021-02-20
Published:2021-10-26
Online:2021-11-12
摘要:
Bt毒素的长期应用使其在害虫抗药性等方面存在生态风险,这一现状促使高特异活性和新功能Bt抗虫资源的积极开发。前期研究发现抗独特型单链抗体制备技术是开发新型杀虫蛋白的一个新途径。然而获得的Bt Cry1B抗独特型单链抗体C7与昆虫BBMV结合能力不高,需进一步改造提高。通过分子模拟技术对C7与稻纵卷叶螟BBMV上的受体氨肽酶N进行同源建模、分子对接,并预测C7与氨肽酶N结合区域的热点残基。利用C7的热点残基构建饱和突变抗体库,并采用固相筛选方法,得到突变体Y124G。BIAcore分析表明Y124G与BBMV的结合能力增加,生物测定发现对稻纵卷叶螟的杀虫活性提高。这为抗体分子改造技术的应用奠定了基础,也为新型生物农药创制提供了新的思路。
仲建锋, 李兴奎, 徐重新, 张霄, 刘贤金. Cry1B抗独特型单链抗体的定点突变及生物活性分析[J]. 生物技术通报, 2021, 37(10): 186-195.
ZHONG Jian-feng, LI Xing-kui, XU Chong-xin, ZHANG Xiao, LIU Xian-jin. Biological Activity of Anti-idiotypic Single Chain Fragment Variable Antibody Against Cry1B by Site-directed Mutagenesis[J]. Biotechnology Bulletin, 2021, 37(10): 186-195.
| 引物名称 Primer | 序列 Sequence(5'-3') |
|---|---|
| LMB3 | CAGGAAACAGCTATGAC |
| pHENseq | CTATGCGGCCCCATTCA |
| NNK_VH | CCCCAGTAGTCAAANNKNNKACCAGATTTCGC |
| NNK_VL | CTCCTGATCTATNNKGCATCCNNKTTGCAAAGTGG |
表1 饱和突变抗体库构建使用的引物序列
Table 1 Primer sequences for the construction of site-directed mutagenesis library
| 引物名称 Primer | 序列 Sequence(5'-3') |
|---|---|
| LMB3 | CAGGAAACAGCTATGAC |
| pHENseq | CTATGCGGCCCCATTCA |
| NNK_VH | CCCCAGTAGTCAAANNKNNKACCAGATTTCGC |
| NNK_VL | CTCCTGATCTATNNKGCATCCNNKTTGCAAAGTGG |
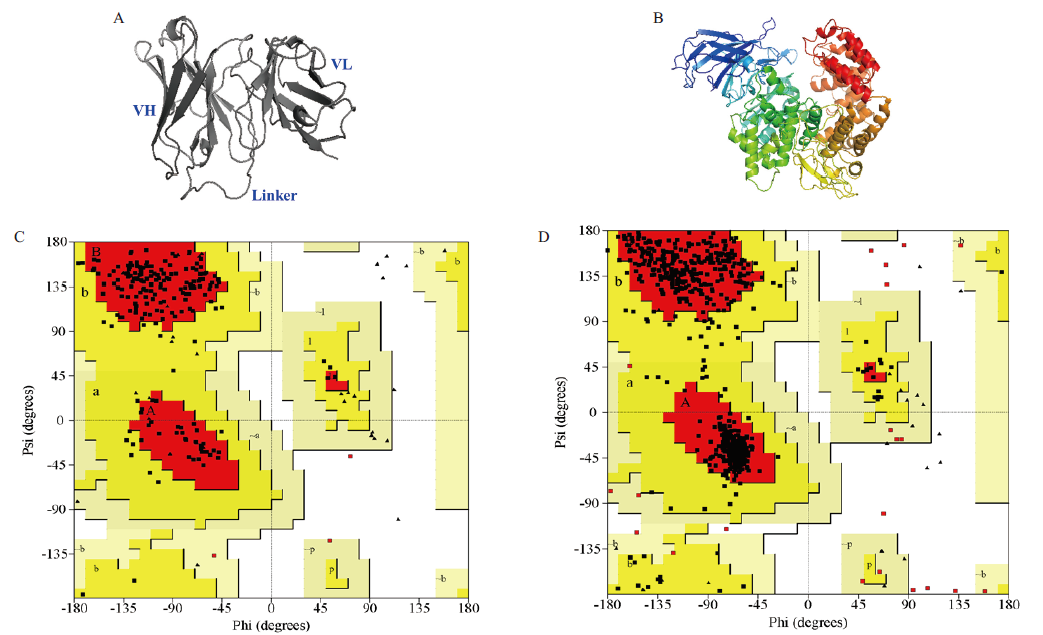
图2 ScFv-C7和CmAPN三维结构的同源建模 A:ScFv-C7的三维结构模拟;B:CmAPN三维结构模拟;C:ScFv-C7的Ramachandran图;D:CmAPN的Ramachandran图
Fig. 2 Three-dimensional structure models of scFv-C7 and CmAPN A:Three-dimensional structure model of scFv-C7. B:3D structure model of CmAPN. C:Ramachandran plot for scFv-C7 model D:Ramachandran plot for CmAPN model
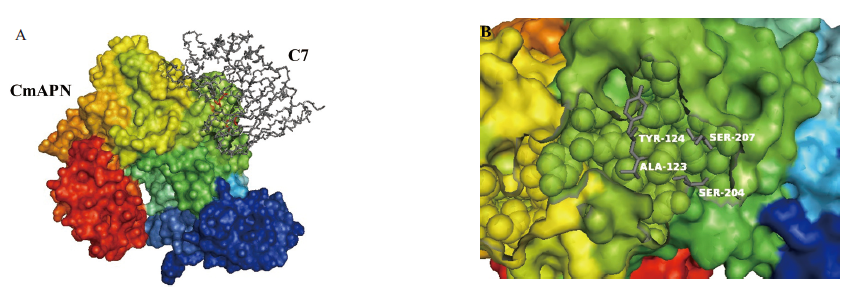
图3 scFv-C7和CmAPN的分子对接及互作热点分析 A:ScFv-C7和CmAPN的分子对接;B:ScFv-C7在接触界面上的热点残基
Fig. 3 Molecular docking and the interaction hot spot analysis between scFv-C7 and CmAPN A:Molecular docking of scFv-C7 and CmAPN. B:Predicted hot spot residues on binding interface from scFv-C7
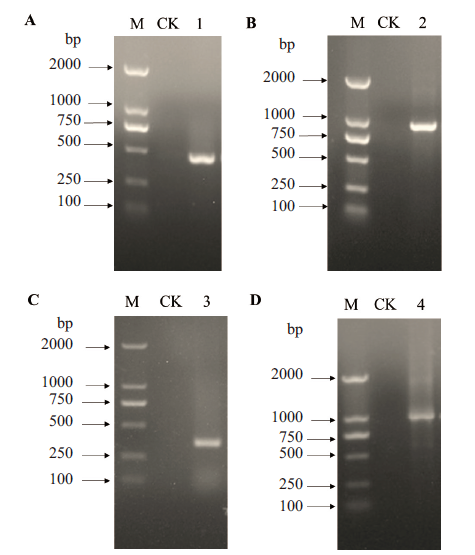
图4 基于scFv-C7对接界面热点氨基酸的饱和突变 A:VH“大引物”的扩增;B:带有H-CDR3饱和突变位点的scFv-C7全长扩增;C:VL“大引物”的扩增;D:带有H-CDR3和L-CDR2饱和突变的scFv-C7全长扩增;M:DNA分子量;1:VH“大引物”;2:带有H-CDR3饱和突变的PCR产物;3:VL“大引物”;4:带有H-CDR3和L-CDR2饱和突变位点的PCR产物;
Fig. 4 Saturation mutagenesis of hot spot amino acids on docking interface based on scFv-C7 A:Amplification of VH mega-primer. B:Full-length amplification of scFv-C7 with H-CDR3 saturation mutagenesis site. C:Amplification of VL mega-primer. D:Full-length amplification of scFv-C7 with H-CDR3 and L-CDR2 saturation mutagenesis sites. M:DNA marker. 1:VH mega-primer. 2:PCR product with H-CDR3 saturation mutagenesis. 3:VL mega-primer. 4:PCR product with H-CDR3 and L-CDR2 saturation mutagenesis sites
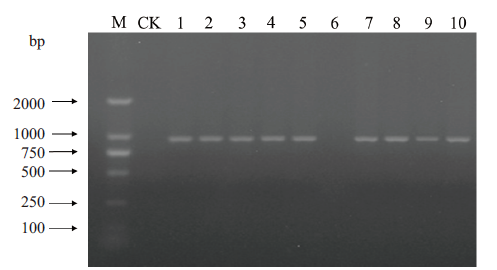
图5 构建的饱和突变抗体库正确性验证 M:DNA分子量;CK:空白对照;1-10:10个随机挑取的单克隆
Fig.5 Verification of correction from constructed satura-tion mutant antibody library M:DNA marker. CK:Blank control. 1-10:Ten pieces of randomly picked monoclones
| 富集轮数 Enriching rounds | 包被蛋白 Coating antigen protein | 包被浓度 Coating antigen concentration /(μg·mL-1) | 投入量 Input | 产出量 Output | 产出/投入 Output/Input |
|---|---|---|---|---|---|
| 1 | BBMV | 100 | 2.0×106 | 6.0×102 | 3.0×10-4 |
| 2 | BBMV | 50 | 1.8×106 | 3.5×103 | 1.9×10-3 |
| 3 | BBMV | 25 | 2.1×106 | 7.5×103 | 3.6×10-3 |
表2 scFv-C7突变抗体库的富集和筛选
Table 2 Enrichment and screening of scFv-C7 mutant antibody library
| 富集轮数 Enriching rounds | 包被蛋白 Coating antigen protein | 包被浓度 Coating antigen concentration /(μg·mL-1) | 投入量 Input | 产出量 Output | 产出/投入 Output/Input |
|---|---|---|---|---|---|
| 1 | BBMV | 100 | 2.0×106 | 6.0×102 | 3.0×10-4 |
| 2 | BBMV | 50 | 1.8×106 | 3.5×103 | 1.9×10-3 |
| 3 | BBMV | 25 | 2.1×106 | 7.5×103 | 3.6×10-3 |
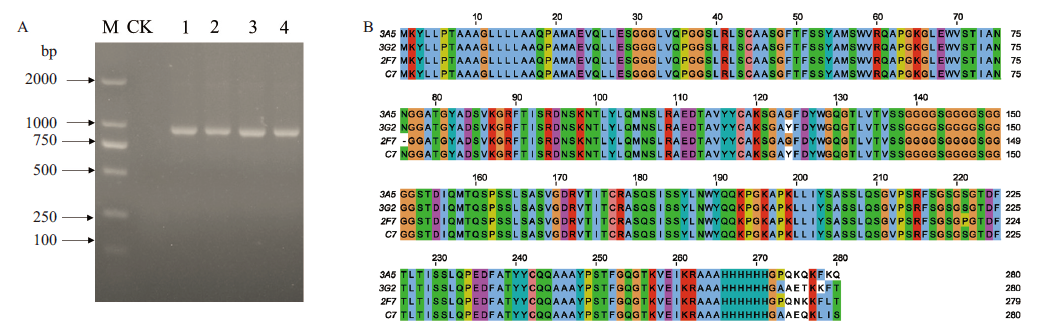
图7 突变单链抗体的基因克隆(A)与氨基酸序列比对(B) M:2 000 bp DNA maker;CK:空白对照;1-4分别表示3A5、3G2、2F7、C7基因
Fig.7 Gene cloning(A)and amino acid sequence alignment(B)of mutant scFv antibodies M:2 000 bp DNA marker. CK:Blank control. 1- 4 refers to 3A5,3G2,2F7 and C7 gene,respectively
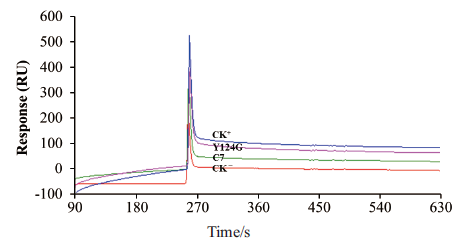
图8 SPR分析突变单链抗体与CmBBMV的结合 CK-:阴性对照scFv-D5;CK+:阳性对照Cry1B毒素
Fig. 8 SPR analysis of mutant scFv antibodies binding to CmBBMV CK-:Negative control scFv-D5. CK+:Positive control(Cry1B toxin)
| 样品 Samples | 校正死亡率Corrected mortality/% | ||
|---|---|---|---|
| 24 h | 48 h | 72 h | |
| CK- | 0.00 ±0.00c | 0.00 ±0.00d | 4.44±1.11d |
| CK+ | 35.56±2.94a | 62.22±1.11a | 76.67±5.09a |
| C7 | 25.56±2.94b | 27.78±1.11c | 42.22±2.94c |
| Y124G | 21.11±2.22b | 34.44±1.11b | 56.67±1.92b |
表3 突变单链抗体对稻纵卷叶螟的生物测定
Table 3 Bioassay of mutant scFv antibodies against C. medinalis larvae
| 样品 Samples | 校正死亡率Corrected mortality/% | ||
|---|---|---|---|
| 24 h | 48 h | 72 h | |
| CK- | 0.00 ±0.00c | 0.00 ±0.00d | 4.44±1.11d |
| CK+ | 35.56±2.94a | 62.22±1.11a | 76.67±5.09a |
| C7 | 25.56±2.94b | 27.78±1.11c | 42.22±2.94c |
| Y124G | 21.11±2.22b | 34.44±1.11b | 56.67±1.92b |
| [1] |
Bravo A, Gómez I, Porta H, et al. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity[J]. Microb Biotechnol, 2013, 6(1):17-26.
doi: 10.1111/j.1751-7915.2012.00342.x URL |
| [2] |
Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity[J]. Microbiol Mol Biol Rev, 2007, 71(2):255-281.
doi: 10.1128/MMBR.00034-06 URL |
| [3] |
Jurat-Fuentes JL, Crickmore N. Specificity determinants for Cry insecticidal proteins:Insights from their mode of action[J]. J Invertebr Pathol, 2017, 142:5-10.
doi: S0022-2011(16)30103-3 pmid: 27480404 |
| [4] | Heckel DG. How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective[J]. Arch Insect Biochem Physiol, 2020, 104(2):e21673. |
| [5] |
Knight PJ, Carroll J, Ellar DJ. Analysis of glycan structures on the 120 kDa aminopeptidase N of Manduca sexta and their interactions with Bacillus thuringiensis Cry1Ac toxin[J]. Insect Biochem Mol Biol, 2004, 34(1):101-112.
doi: 10.1016/j.ibmb.2003.09.007 URL |
| [6] |
Luo K, Sangadala S, Masson L, et al. The Heliothis virescens 170 kDa aminopeptidase functions as “Receptor A” by mediating specific Bacillus thuringiensis Cry1A δ-endotoxin binding and pore formation[J]. Insect Biochem Mol Biol, 1997, 27(8/9):735-743.
doi: 10.1016/S0965-1748(97)00052-0 URL |
| [7] |
Simpson RM, Newcomb RD. Binding of Bacillus thuringiensis delta-endotoxins Cry1Ac and Cry1Ba to a 120-kDa aminopeptidase-N of Epiphyas postvittana purified from both brush border membrane vesicles and baculovirus-infected Sf9 cells[J]. Insect Biochem Mol Biol, 2000, 30(11):1069-1078.
doi: 10.1016/S0965-1748(00)00082-5 URL |
| [8] | 刘贤金, 徐重新, 张霄, 等. 一种人源抗虫基因及其编码的抗Cry1B毒素独特型单链抗体与应用:中国,201410037175.X[P]. 2015-11-18. |
| Liu XJ, Xu CX, Zhang X, et al. The anti-Cry1B toxin idiotype single-chain antibody of a kind of people source anti insect gene and coding thereof and application: China, 201410037175. X[P]. 2015-11-18. | |
| [9] |
Bird RE, Walker BW. Single chain antibody variable regions[J]. Trends Biotechnol, 1991, 9(1):132-137.
doi: 10.1016/0167-7799(91)90044-I URL |
| [10] |
Perchiacca JM, Tessier PM. Engineering aggregation-resistant antibodies[J]. Annu Rev Chem Biomol Eng, 2012, 3(1):263-286.
doi: 10.1146/chembioeng.2012.3.issue-1 URL |
| [11] |
Qiao C, Lv M, Li X, et al. Affinity maturation of antiHER2 monoclonal antibody MIL5 using an epitope-specific synthetic phage library by computational design[J]. J Biomol Struct Dyn, 2013, 31(5):511-521.
doi: 10.1080/07391102.2012.706073 URL |
| [12] |
Sheedy C, Roger MacKenzie C, Hall JC. Isolation and affinity maturation of hapten-specific antibodies[J]. Biotechnol Adv, 2007, 25(4):333-352.
doi: 10.1016/j.biotechadv.2007.02.003 URL |
| [13] |
Barderas R, Desmet J, Timmerman P, et al. Affinity maturation of antibodies assisted by in silico modeling[J]. PNAS, 2008, 105(26):9029-9034.
doi: 10.1073/pnas.0801221105 pmid: 18574150 |
| [14] |
Zhang J, Valianou M, Simmons H, et al. Identification of inhibitory scFv antibodies targeting fibroblast activation protein utilizing phage display functional screens[J]. Faseb J, 2013, 27(2):581-589.
doi: 10.1096/fsb2.v27.2 URL |
| [15] |
Kuntal BK, Aparoy P, Reddanna P. EasyModeller:a graphical interface to MODELLER[J]. BMC Res Notes, 2010, 3:226.
doi: 10.1186/1756-0500-3-226 URL |
| [16] | Schrödingerllc. The PyMOL molecular graphics system, Version 1. 3r1[M]. Portland, Oregon, USA: Schrödinger, LLC, 2010. |
| [17] |
Barik S. Mutagenesis and gene fusion by megaprimer PCR[J]. Methods Mol Biol, 1997, 67:173-182.
pmid: 9031141 |
| [18] |
Séraphin B, Kandels-Lewis S. An efficient PCR mutagenesis strategy without gel purificiation step that is amenable to automation[J]. Nucleic Acids Res, 1996, 24(16):3276-3277.
pmid: 8774913 |
| [19] |
Wolfersberger M, Luethy P, Maurer A, et al. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly(Pieris brassicae)[J]. Comp Biochem Physiol Part A:Physiol, 1987, 86(2):301-308.
doi: 10.1016/0300-9629(87)90334-3 URL |
| [20] | 焦凌霞, 徐茜茜, 刘媛, 等. 人源化抗苏云金芽孢杆菌Cry1Ab毒素单域抗体的筛选及活性鉴定[J]. 中国食品学报, 2017, 17(10):268-273. |
| Jiao LX, Xu XX, Liu Y, et al. Screening and identification of humanized single domain antibodies(sdAbs)against Bacillus thuringiensis Cry1Ab toxin[J]. J Chin Inst Food Sci Technol, 2017, 17(10):268-273. | |
| [21] |
Malia TJ, Obmolova G, Almagro JC, et al. Crystal structure of human germline antibody 3-23/B3[J]. Mol Immunol, 2011, 48(12/13):1586-1588.
doi: 10.1016/j.molimm.2011.04.020 URL |
| [22] |
James LC, Jones PC, McCoy A, et al. Β-edge interactions in a pentadecameric human antibody vκ domain[J]. J Mol Biol, 2007, 367(3):603-608.
pmid: 17292396 |
| [23] |
Wong AH, Zhou D, Rini JM. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing[J]. J Biol Chem, 2012, 287(44):36804-36813.
doi: 10.1074/jbc.M112.398842 URL |
| [24] |
Lippow SM, Wittrup KD, Tidor B. Computational design of antibody-affinity improvement beyond in vivo maturation[J]. Nat Biotechnol, 2007, 25(10):1171-1176.
pmid: 17891135 |
| [25] |
Clark LA, Boriack-Sjodin PA, Eldredge J, et al. Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design[J]. Protein Sci, 2006, 15(5):949-960.
doi: 10.1110/ps.052030506 URL |
| [26] |
Wen K, Nölke G, Schillberg S, et al. Improved fluoroquinolone detection in ELISA through engineering of a broad-specific single-chain variable fragment binding simultaneously to 20 fluoroquinolones[J]. Anal Bioanal Chem, 2012, 403(9):2771-2783.
doi: 10.1007/s00216-012-6062-z URL |
| [27] |
Tian J, Wang P, Gao S, et al. Enhanced thermostability of methyl parathion hydrolase from Ochrobactrum sp. M231 by rational engineering of a Glycine to proline mutation[J]. Febs J, 2010, 277(23):4901-4908.
doi: 10.1111/j.1742-4658.2010.07895.x URL |
| [28] |
Boozer C, Kim G, Cong SX, et al. Looking towards label-free biomolecular interaction analysis in a high-throughput format:a review of new surface plasmon resonance technologies[J]. Curr Opin Biotechnol, 2006, 17(4):400-405.
doi: 10.1016/j.copbio.2006.06.012 URL |
| [29] |
McDonnell JM. Surface plasmon resonance:towards an understanding of the mechanisms of biological molecular recognition[J]. Curr Opin Chem Biol, 2001, 5(5):572-577.
pmid: 11578932 |
| [30] |
Homola J, Yee SS, Gauglitz G. Surface plasmon resonance sensors:review[J]. Sensor Actuat B:Chem, 1999, 54(1/2):3-15.
doi: 10.1016/S0925-4005(98)00321-9 URL |
| [31] |
Zhuang X, Stahl SJ, Watts NR, et al. A cell-penetrating antibody fragment against HIV-1 Rev has high antiviral activity:characterization of the paratope[J]. J Biol Chem, 2014, 289(29):20222-20233.
doi: 10.1074/jbc.M114.581090 URL |
| [32] |
Richter A, Eggenstein E, Skerra A. Anticalins:exploiting a non-Ig scaffold with hypervariable loops for the engineering of binding proteins[J]. FEBS Lett, 2014, 588(2):213-218.
doi: 10.1016/j.febslet.2013.11.006 pmid: 24239535 |
| [33] | 李敏, 郭美琪, 相伟芳, 等. 分子对接技术在昆虫化学感受研究中的应用进展[J]. 植物保护, 2019, 45(5):121-127. |
| Li M, Guo MQ, Xiang WF, et al. Research progress in molecular docking in insect chemosense[J]. Plant Prot, 2019, 45(5):121-127. |
| [1] | 韩惠, 张舰, 任宇红. 短链脱氢酶Lvchun的分子改造及其在氯霉胺合成中的应用[J]. 生物技术通报, 2023, 39(4): 81-92. |
| [2] | 朱秋雨, 段绪果. L-天冬氨酸-α-脱羧酶的重组表达、定点突变及高通量检测方法的建立[J]. 生物技术通报, 2022, 38(5): 269-278. |
| [3] | 田庚, 高伟强, 陈晓波, 张春晓. 地衣芽孢杆菌KD-1β-甘露聚糖酶定点突变提高酶活性及稳定性[J]. 生物技术通报, 2021, 37(10): 100-109. |
| [4] | 孙熙麟, 蒋振彦, 刘志屹, 戴璐, 孙非, 黄伟. 氨基酸定点突变提高灵芝蛋白LZ-8热稳定性的研究[J]. 生物技术通报, 2020, 36(1): 23-28. |
| [5] | 邱锦, 黄火清, 姚斌, 罗会颖. 解淀粉芽孢杆菌淀粉酶催化活力改良及其在枯草芽孢杆菌中的高效表达[J]. 生物技术通报, 2019, 35(9): 134-143. |
| [6] | 徐林娜, 胡孟可, 童文艳, 李芬. 烟草NtTkr尾部点突变对卷曲螺旋结构及与靶蛋白相互作用的影响[J]. 生物技术通报, 2019, 35(5): 64-69. |
| [7] | 于爽, 李帅, 李海涛, 刘荣梅, 高继国. 苏云金芽胞杆菌Vip3AaAdAa嵌合蛋白的构建及其杀虫活性分析[J]. 生物技术通报, 2019, 35(4): 51-56. |
| [8] | 王柳月, 李慧美, 马梦琪, 梁明星, 贺如阳, 陈华波. 利用旁侧引物提高重叠延伸PCR定点突变效率[J]. 生物技术通报, 2019, 35(12): 196-202. |
| [9] | 华晨, 李新新, 涂涛, 杨虹, 罗会颖, 陈家明, 姚斌, 柏映国, 彭书传. 基于酶热稳定性系统计算的乳酸氧化酶热稳定性改造[J]. 生物技术通报, 2018, 34(8): 144-150. |
| [10] | 陈少威, 吴程, 苏月华, 蔡斌斌, 谢盼盼, 杨梅. 苏云金芽胞杆菌aiiA的5'端侧翼序列的克隆与功能鉴定[J]. 生物技术通报, 2018, 34(11): 136-143. |
| [11] | 秦海彬, 熊涛, 张博, 牛坤. α-酮戊二酸半醛脱氢酶的定点突变及酶学性质变化 [J]. 生物技术通报, 2017, 33(8): 180-185. |
| [12] | 曾静, 郭建军, 袁林, 杨罡, 陈俊. 极端嗜热α-淀粉酶ApkA的高温活性和热稳定性的优化研究[J]. 生物技术通报, 2017, 33(8): 192-198. |
| [13] | 张月李海涛刘荣梅高继国. 苏云金芽胞杆菌cry2Ab34基因的克隆、表达和杀虫活性分析[J]. 生物技术通报, 2017, 33(4): 185-190. |
| [14] | 徐曼, 蒋健, 束长龙, 张杰, 宋福平. Cry1Ab/Cry1Ah杂合蛋白构建与功能研究[J]. 生物技术通报, 2015, 31(9): 91-96. |
| [15] | 刘松,陆信曜,周景文,堵国成,陈坚. 脂肪氧合酶结构、分子改造与发酵研究进展[J]. 生物技术通报, 2015, 31(12): 34-41. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||