








生物技术通报 ›› 2021, Vol. 37 ›› Issue (7): 98-106.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0628
薛清1( ), 杜虹锐1, 薛会英2, 王译浩1, 王暄1(
), 杜虹锐1, 薛会英2, 王译浩1, 王暄1( ), 李红梅1
), 李红梅1
收稿日期:2021-06-28
出版日期:2021-07-26
发布日期:2021-08-13
作者简介:薛清,博士,副教授,研究方向:植物线虫学;E-mail:基金资助:
XUE Qing1( ), DU Hong-rui1, XUE Hui-ying2, WANG Yi-hao1, WANG Xuan1(
), DU Hong-rui1, XUE Hui-ying2, WANG Yi-hao1, WANG Xuan1( ), LI Hong-mei1
), LI Hong-mei1
Received:2021-06-28
Published:2021-07-26
Online:2021-08-13
摘要:
苜蓿滑刃线虫(Aphelenchoides medicagus)是一种兼性植物寄生线虫,可在多种真菌上完成生活史,同时对大豆和苜蓿具有弱寄生性。本研究采用Illumina平台对苜蓿滑刃线虫进行低覆盖全基因组测序,从获得的全基因组序列中提取线粒体基因,并利用“种子”序列进行组装;同时对其中富含AT的非编码部分进行PCR扩增与Sanger测序,将扩增与组装的结果进行拼接,共获得14 411 bp的线粒体基因组。基因注释结果显示,苜蓿滑刃线虫共有蛋白编码基因12个,tRNA基因22个,rRNA基因2个,其基因构成和排列与贝西滑刃线虫(A. besseyi)、松材线虫(Bursaphelenchus xylophilus)及拟松材线虫(B. mucronatus)完全相同。基于氨基酸序列的系统发育显示,苜蓿滑刃线虫与贝西滑刃线虫互为姐妹群,且与松材线虫、拟松材线虫处在高度支持的滑刃线虫科单系中。本研究表明,低覆盖全基因组测序法可以成功组装出线粒体基因组的大部分序列,证明了利用该方法获取线虫线粒体全基因组序列的可行性。
薛清, 杜虹锐, 薛会英, 王译浩, 王暄, 李红梅. 苜蓿滑刃线虫线粒体基因组及其系统发育研究[J]. 生物技术通报, 2021, 37(7): 98-106.
XUE Qing, DU Hong-rui, XUE Hui-ying, WANG Yi-hao, WANG Xuan, LI Hong-mei. Mitochondrial Genome and Phylogeny of Aphelenchoides medicagus[J]. Biotechnology Bulletin, 2021, 37(7): 98-106.
| 基因名 Gene name | 起始位点 Start | 终止位点 Stop | 长度 Length/bp | 基因间隔 Intergenic space | 起始/终止密码子 Start/stop codons |
|---|---|---|---|---|---|
| cox1 | 955 | 2535 | 1 581 | 0 | ATT/TAA |
| trnC(gca) | 2534 | 2587 | 54 | -2 | |
| trnM(cat) | 2588 | 2641 | 54 | 0 | |
| trnD(gtc) | 2642 | 2696 | 55 | 0 | |
| trnG(tcc) | 2697 | 2751 | 55 | 0 | |
| cox2 | 2761 | 3450 | 690 | 9 | ATT/TAA |
| trnH(gtg) | 3450 | 3503 | 54 | -1 | |
| rrnL | 3505 | 4415 | 911 | 1 | |
| nad3 | 4423 | 4758 | 336 | 7 | ATA/TAA |
| nad5 | 4842 | 6347 | 1 506 | 83 | ATT/TAG |
| trnA(tgc) | 6346 | 6399 | 54 | -2 | |
| trnP(tgg) | 6400 | 6453 | 54 | 0 | |
| trnV(tac) | 6453 | 6508 | 56 | -1 | |
| nad6 | 6521 | 6949 | 429 | 12 | ATT/TAA |
| nad4L | 6952 | 7180 | 229 | 2 | TTG/T |
| trnW(tca) | 7181 | 7236 | 56 | 0 | |
| trnE(ttc) | 7243 | 7296 | 54 | 6 | |
| rrnS | 7297 | 7963 | 667 | 0 | |
| trnS2(tga) | 7966 | 8019 | 54 | 2 | |
| trnY(gta) | 8021 | 8074 | 54 | 1 | |
| nad1 | 8066 | 8944 | 879 | -9 | ATG/TAA |
| atp6 | 8944 | 9543 | 600 | -1 | ATT/TAA |
| trnK(ttt) | 9542 | 9598 | 57 | -2 | |
| trnL2(taa) | 9599 | 9653 | 55 | 0 | |
| trnS1(tct) | 9654 | 9706 | 53 | 0 | |
| nad2 | 9720 | 10536 | 817 | 13 | ATA/T |
| trnI(gat) | 10546 | 10604 | 59 | 9 | |
| trnR(acg) | 10601 | 10654 | 54 | -4 | |
| trnQ(ttg) | 10655 | 10708 | 54 | 0 | |
| trnF(gaa) | 10709 | 10762 | 54 | 0 | |
| cob | 10760 | 11884 | 1 125 | -3 | ATA/TAA |
| trnL1(tag) | 11880 | 11936 | 57 | -5 | |
| cox3 | 11942 | 12691 | 750 | 5 | ATT/TAG |
| trnN(gtt) | 12692 | 12745 | 54 | 0 | |
| trnT(tgt) | 12746 | 12800 | 55 | 0 | |
| nad4 | 12807 | 14033 | 1 227 | 6 | ATA/TAA |
表1 苜蓿滑刃线虫的线粒体基因位置分布与起始终止密码子
Table 1 Placements and start/stop codons of mitochondrial genes in A. medicagus
| 基因名 Gene name | 起始位点 Start | 终止位点 Stop | 长度 Length/bp | 基因间隔 Intergenic space | 起始/终止密码子 Start/stop codons |
|---|---|---|---|---|---|
| cox1 | 955 | 2535 | 1 581 | 0 | ATT/TAA |
| trnC(gca) | 2534 | 2587 | 54 | -2 | |
| trnM(cat) | 2588 | 2641 | 54 | 0 | |
| trnD(gtc) | 2642 | 2696 | 55 | 0 | |
| trnG(tcc) | 2697 | 2751 | 55 | 0 | |
| cox2 | 2761 | 3450 | 690 | 9 | ATT/TAA |
| trnH(gtg) | 3450 | 3503 | 54 | -1 | |
| rrnL | 3505 | 4415 | 911 | 1 | |
| nad3 | 4423 | 4758 | 336 | 7 | ATA/TAA |
| nad5 | 4842 | 6347 | 1 506 | 83 | ATT/TAG |
| trnA(tgc) | 6346 | 6399 | 54 | -2 | |
| trnP(tgg) | 6400 | 6453 | 54 | 0 | |
| trnV(tac) | 6453 | 6508 | 56 | -1 | |
| nad6 | 6521 | 6949 | 429 | 12 | ATT/TAA |
| nad4L | 6952 | 7180 | 229 | 2 | TTG/T |
| trnW(tca) | 7181 | 7236 | 56 | 0 | |
| trnE(ttc) | 7243 | 7296 | 54 | 6 | |
| rrnS | 7297 | 7963 | 667 | 0 | |
| trnS2(tga) | 7966 | 8019 | 54 | 2 | |
| trnY(gta) | 8021 | 8074 | 54 | 1 | |
| nad1 | 8066 | 8944 | 879 | -9 | ATG/TAA |
| atp6 | 8944 | 9543 | 600 | -1 | ATT/TAA |
| trnK(ttt) | 9542 | 9598 | 57 | -2 | |
| trnL2(taa) | 9599 | 9653 | 55 | 0 | |
| trnS1(tct) | 9654 | 9706 | 53 | 0 | |
| nad2 | 9720 | 10536 | 817 | 13 | ATA/T |
| trnI(gat) | 10546 | 10604 | 59 | 9 | |
| trnR(acg) | 10601 | 10654 | 54 | -4 | |
| trnQ(ttg) | 10655 | 10708 | 54 | 0 | |
| trnF(gaa) | 10709 | 10762 | 54 | 0 | |
| cob | 10760 | 11884 | 1 125 | -3 | ATA/TAA |
| trnL1(tag) | 11880 | 11936 | 57 | -5 | |
| cox3 | 11942 | 12691 | 750 | 5 | ATT/TAG |
| trnN(gtt) | 12692 | 12745 | 54 | 0 | |
| trnT(tgt) | 12746 | 12800 | 55 | 0 | |
| nad4 | 12807 | 14033 | 1 227 | 6 | ATA/TAA |
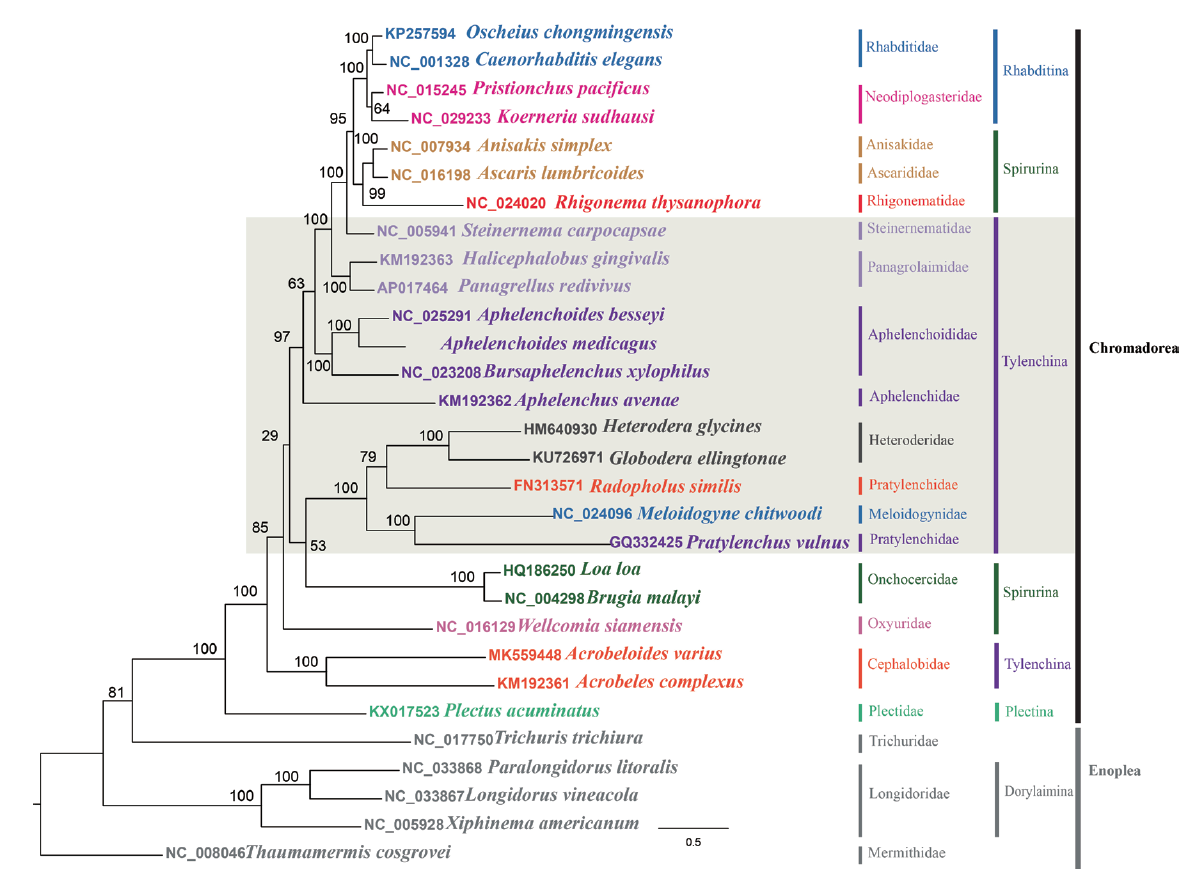
图4 基于12个蛋白编码基因的氨基酸序列构建的极大似然系统发育树
Fig. 4 Phylogenetic tree based on amino acid sequences from concatenated 12 protein coding genes by maximum likelihood method
| [1] |
Sánchez-Monge A,Flores L,Salazar L,et al.An updated list of the plants associated with plant-parasitic Aphelenchoides(Nematoda:Aphelenchoididae)and its implications for plant-parasitism within this genus[J].Zootaxa,2015,4013(2):207-224.
doi: 10.11646/zootaxa.4013.2.3 pmid: 26623893 |
| [2] | Hunt DJ.Aphelenchida, Longidoridae and Trichodoridae:Their Systematics and Bionomics[M].UK:CAB International, Wallingford,1993. |
| [3] |
McCuiston JL,Hudson LC,Subbotin SA,et al.Conventional and PCR detection of Aphelenchoides fragariae in diverse ornamental host plant species[J].J Nematol,2007,39(4):343-355.
pmid: 19259510 |
| [4] |
Jones JT,Haegeman A,Danchin EGJ,et al.Top 10 plant-parasitic nematodes in molecular plant pathology[J].Mol Plant Pathol,2013,14(9):946-961.
doi: 10.1111/mpp.12057 pmid: 23809086 |
| [5] |
Wang Z,Bert W,Gu JF,et al.Aphelenchoides medicagus n. sp. (Tylenchina:Aphelenchoididae)found in Medicago sativa imported into China from the USA[J].Nematology,2019,21(7):709-723.
doi: 10.1163/15685411-00003247 URL |
| [6] | 王珍,李冉,李红梅,等.不同真菌和植物种类对苜蓿滑刃线虫繁殖的影响[J].植物保护,2021,47(1):97-102. |
| Wang Z,Li R,Li HM,et al.Effects of different fungi and plant species on reproduction of Aphelenchoides medicagus[J].Plant Prot,2021,47(1):97-102. | |
| [7] |
Holterman M,Holovachov O,van den Elsen S,et al.Small subunit ribosomal DNA-based phylogeny of basal Chromadoria(Nematoda)suggests that transitions from marine to terrestrial habitats(and vice versa)require relatively simple adaptations[J].Mol Phylogenetics Evol,2008,48(2):758-763.
doi: 10.1016/j.ympev.2008.04.033 URL |
| [8] |
Sultana T,Kim J,Lee SH,et al.Comparative analysis of complete mitochondrial genome sequences confirms independent origins of plant-parasitic nematodes[J].BMC Evol Biol,2013,13:12.
doi: 10.1186/1471-2148-13-12 pmid: 23331769 |
| [9] |
Hegedusova E,Brejova B,Tomaska L,et al.Mitochondrial genome of the basidiomycetous yeast Jaminaea angkorensis[J].Curr Genet,2014,60(1):49-59.
doi: 10.1007/s00294-013-0410-1 pmid: 24071901 |
| [10] |
Kern EMA,Kim T,Park JK.The mitochondrial genome in nematode phylogenetics[J].Front Ecol Evol,2020,8:250. DOI:10.3389/fevo,2020.00250.
doi: 10.3389/fevo,2020.00250 URL |
| [11] |
Sun LH,Zhuo K,Wang HH,et al.The complete mitochondrial genome of Aphelenchoides besseyi(Nematoda:Aphelenchoididae), the first sequenced representative of the subfamily Aphelenchoidinae[J].Nematology,2014,16(10):1167-1180.
doi: 10.1163/15685411-00002844 URL |
| [12] |
Humphreys-Pereira DA,Elling AA.Mitochondrial genomes of Meloidogyne chitwoodi and M. incognita(Nematoda:Tylenchina):comparative analysis, gene order and phylogenetic relationships with other nematodes[J].Mol Biochem Parasitol,2014,194(1/2):20-32.
doi: 10.1016/j.molbiopara.2014.04.003 URL |
| [13] |
Kim T,Lee Y,Kil HJ,et al.The mitochondrial genome of Acrobeloides varius(Cephalobomorpha)confirms non-monophyly of Tylenchina(Nematoda)[J].PeerJ,2020,8:e9108.
doi: 10.7717/peerj.9108 URL |
| [14] |
Ye F,Samuels DC,Clark T,et al.High-throughput sequencing in mitochondrial DNA research[J].Mitochondrion,2014,17:157-163.
doi: 10.1016/j.mito.2014.05.004 URL |
| [15] |
Phillips WS,Brown AMV,Howe DK,et al.The mitochondrial genome of Globodera ellingtonae is composed of two circles with segregated gene content and differential copy numbers[J].BMC Genom,2016,17:706.
doi: 10.1186/s12864-016-3047-x URL |
| [16] |
Ma XY,Agudelo P,Richards VP,et al.The complete mitochondrial genome of the Columbia lance nematode, Hoplolaimus columbus, a major agricultural pathogen in North America[J].Parasites Vectors,2020,13(1):321.
doi: 10.1186/s13071-020-04187-y URL |
| [17] |
Kanzaki N,Futai K.A PCR primer set for determination of phylogenetic relationships of Bursaphelenchus species within the Xylophilus group[J].Nematology,2002,4(1):35-41.
doi: 10.1163/156854102760082186 URL |
| [18] |
Chen S,Zhou Y,Chen Y,et al.Fastp:an ultra-fast all-in-one FASTQ preprocessor[J].Bioinformatics,2018,34(17):i884-i890.
doi: 10.1093/bioinformatics/bty560 URL |
| [19] |
Sedlazeck FJ,Rescheneder P,von Haeseler A.NextGenMap:fast and accurate read mapping in highly polymorphic genomes[J].Bioinformatics,2013,29(21):2790-2791.
doi: 10.1093/bioinformatics/btt468 pmid: 23975764 |
| [20] |
Li H,Handsaker B,Wysoker A,et al.The sequence alignment/map format and SAMtools[J].Bioinformatics,2009,25(16):2078-2079.
doi: 10.1093/bioinformatics/btp352 URL |
| [21] |
Meng G,Li Y,Yang C,et al.MitoZ:a toolkit for animal mitochondrial genome assembly, annotation and visualization[J].Nucleic Acids Res,2019,47(11):e63.
doi: 10.1093/nar/gkz173 URL |
| [22] |
Bernt M,Donath A,Jühling F,et al.MITOS:improved de novo metazoan mitochondrial genome annotation[J].Mol Phylogenet Evol,2013,69(2):313-319.
doi: 10.1016/j.ympev.2012.08.023 URL |
| [23] |
Jühling F,Pütz J,Bernt M,et al.Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements[J].Nucleic Acids Res,2012,40(7):2833-2845.
doi: 10.1093/nar/gkr1131 pmid: 22139921 |
| [24] |
Kerpedjiev P,Hammer S,Hofacker IL.Forna(force-directed RNA):simple and effective online RNA secondary structure diagrams[J].Bioinformatics,2015,31(20):3377-3379.
doi: 10.1093/bioinformatics/btv372 pmid: 26099263 |
| [25] |
Kuraku S,Zmasek CM,Nishimura O,et al.aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity[J].Nucleic Acids Res,2013,41(web server issue):W22-W28.
doi: 10.1093/nar/gkt389 URL |
| [26] |
Lefort V,Longueville JE,Gascuel O.SMS:smart model selection in PhyML[J].Mol Biol Evol,2017,34(9):2422-2424.
doi: 10.1093/molbev/msx149 URL |
| [27] | Miller MA,Pfeiffer W,Schwartz T.Creating the CIPRES Science Gateway for inference of large phylogenetic trees[C]//2010 Gateway Computing Environments Workshop(GCE). NJ:IEEE,2010:1-8. |
| [28] |
Stamatakis A.RAxML version 8:a tool for phylogenetic analysis and post-analysis of large phylogenies[J].Bioinformatics,2014,30(9):1312-1313.
doi: 10.1093/bioinformatics/btu033 URL |
| [29] |
Zhang ZQ.Animal biodiversity:an outline of higher-level classification and survey of taxonomic richness(COVER)[J].Zootaxa,2011,3148(1):3.
doi: 10.11646/zootaxa.3148.1 URL |
| [30] |
Bert W,Leliaert F,Vierstraete AR,et al.Molecular phylogeny of the Tylenchina and evolution of the female gonoduct(Nematoda:Rhabditida)[J].Mol Phylogenet Evol,2008,48(2):728-744.
doi: 10.1016/j.ympev.2008.04.011 pmid: 18502668 |
| [31] |
Gómez-Rodríguez C,Crampton-Platt A,Timmermans MJTN,et al.Validating the power of mitochondrial metagenomics for community ecology and phylogenetics of complex assemblages[J].Methods Ecol Evol,2015,6(8):883-894.
doi: 10.1111/mee3.2015.6.issue-8 URL |
| [32] |
Crampton-Platt A,Timmermans MJ,Gimmel ML,et al.Soup to tree:the phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample[J].Mol Biol Evol,2015,32(9):2302-2316.
doi: 10.1093/molbev/msv111 pmid: 25957318 |
| [1] | 曲春娟, 朱悦, 江晨, 曲明静, 王向誉, 李晓. 铜绿丽金龟线粒体全基因组及其系统发育分析[J]. 生物技术通报, 2023, 39(2): 263-273. |
| [2] | 王平, 丛玲, 王春语, 朱振兴, AAshokKumar, 张丽霞, 陆晓春. 高粱A1型细胞质雄性不育系与保持系线粒体基因组分析比较[J]. 生物技术通报, 2019, 35(5): 42-47. |
| [3] | 金巧, 刘霞, 甘志凯. 木通红喀木虱线粒体基因组的测定与序列分析[J]. 生物技术通报, 2018, 34(11): 127-135. |
| [4] | 董传举, 张松皓, 陈坤慈, 宋迎楠, 徐鹏, 孙效文. 一种乌鳢与斑鳢线粒体PCR-RFLP鉴定方法[J]. 生物技术通报, 2014, 0(8): 59-64. |
| [5] | 魏峦峦;沈斌;沈和定;陈诚;方磊;吴文健;. 一种石磺科贝类线粒体基因组全序列分析[J]. , 2010, 0(08): 188-194. |
| [6] | 杨琴玲;李思发;徐嘉伟;陈琴;王成辉;. 鳙的线粒体基因组核苷酸全序列分析(英文)[J]. , 2009, 0(02): 112-117. |
| [7] | 廖祥儒;朱新产;万怡震. 核酸分析技术与植物的分子分类[J]. , 1995, 0(06): 3-5. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||