








生物技术通报 ›› 2024, Vol. 40 ›› Issue (1): 100-112.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0509
收稿日期:2023-05-30
出版日期:2024-01-26
发布日期:2024-02-06
通讯作者:
张彭湃,男,博士,副教授,研究方向:食品发酵;E-mail: bio_apai@163.com作者简介:王俊芳,女,硕士,高级实验师,研究方向:资源微生物应用;E-mail: jfw188@163.com
基金资助:
WANG Jun-fang( ), HUANG Qiu-bin, ZHANG Piao-dan, ZHANG Peng-pai(
), HUANG Qiu-bin, ZHANG Piao-dan, ZHANG Peng-pai( )
)
Received:2023-05-30
Published:2024-01-26
Online:2024-02-06
摘要:
Surfactin是由多种芽孢杆菌产生的一种脂肽,其由一个环七肽头基通过内脂键与链长12-17个碳原子的β-羟基脂肪酸连接组成,是一种非常有效的生物表面活性剂,并具有重要的生物学功能。本文介绍了surfactin家族的结构和生物合成模式,surfactin是由非核糖体肽合成酶催化合成,这种机制赋予了surfactin家族成员的结构多样性;综述了surfactin在植物病害生防中的作用及机制。已有的研究表明surfactin在生物防治中的作用机制主要有以下4种方式:(1)损伤病原菌细胞膜,引起细胞膜裂解或渗透压失衡;(2)抑制病原体繁殖;(3)诱导植株系统抗性;(4)促进生防菌株的定殖或生物膜的形成。进一步总结了利用基因工程技术研究surfactin的最新进展,以指导surfactin的生物合成及其新衍生物开发。Surfactin的高度结构多样性,使其具有丰富的物理化学特性,这些特性可以与多种生物活性联系在一起,将在不同的领域有更广泛应用。随着对surfactin生物合成研究的日益深入,将会有更多高性能和广阔应用前景的新型脂肽被研发,这为surfactin的进一步研究和生防应用提供依据。
王俊芳, 黄秋斌, 张飘丹, 张彭湃. Surfactin的结构、生物合成及其在生物防治中的作用[J]. 生物技术通报, 2024, 40(1): 100-112.
WANG Jun-fang, HUANG Qiu-bin, ZHANG Piao-dan, ZHANG Peng-pai. Structure and Biosynthesis of Surfactin as well as Its Role in Biological Control[J]. Biotechnology Bulletin, 2024, 40(1): 100-112.

图2 非核糖体肽生物合成的原理 (A)非核糖体肽合成酶(nonribosomal peptide synthetases, NRPSs)相关结构域的晶体结构;(B)NRPSs的作用原理图;(C)NRPSs结构域的催化反应。由TE酶结构域负责脂肽的环化和释放,I为水解方式,得到线性产物;II为环化方式,得到环状产物;具体哪种方式取决于NRPSs模板的识别特性
Fig. 2 Principle of non-ribosomal peptide(NRP)biosynthesis (A)Crystal structure of non-ribosomal peptide synthetases(NRPSs)related domain.(B)The principle of action of NRPSs.(C)Catalytic reaction of NRPSs structure domain. The TE enzyme domain is responsible for the cyclization and release of lipopeptides. In the figure, I is the linear product obtained through hydrolysis method, while II is the cyclization method to obtain cyclic products. The specific method depends on the recognition characteristics of the NRPSs
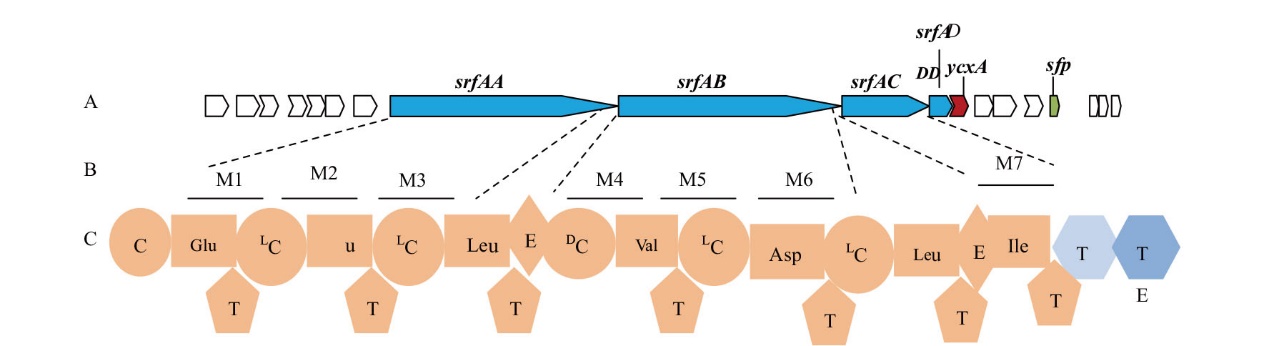
图3 Surfactin合成基因簇组成 A:Surfactin生物合成基因簇,srfAA、srfAB、srfAC和srfAD为NRPS的结构基因;B:SrfAA-SrfAD的经典模块组成;C:SrfAA-SrfAD的结构域
Fig. 3 Composition of gene cluster of surfactin biosynthesis A: Surfactin biosynthesis gene cluster, and the structural genes of NRPS for biosynthesis of surfactin are sfAA, srfAB, srfAC, and srfAD. B: Classic module composition of SrfAA-SrfAD. C: Domain architecture of SrfAA-SrfAD
| [1] |
Li Y, Zou AH, Ye RQ, et al. Effects of molecular structure on surfactin micellization activity[J]. Acta Phys Chim Sin, 2011, 27(5): 1128-1134.
doi: 10.3866/PKU.WHXB20110436 URL |
| [2] |
Kundu D, Hazra C, Chatterjee A, et al. Surfactin-functionalized poly(methyl methacrylate)as an eco-friendly nano-adsorbent: from size-controlled scalable fabrication to adsorptive removal of inorganic and organic pollutants[J]. RSC Adv, 2016, 6(84): 80438-80454.
doi: 10.1039/C6RA10804K URL |
| [3] |
Kim SD, Park SK, Cho JY, et al. Surfactin C inhibits platelet aggregation[J]. J Pharm Pharmacol, 2010, 58(6): 867-870.
doi: 10.1211/jpp.58.6.0018 URL |
| [4] | Jacques P. Surfactin and other lipopeptides from Bacillus spp.[M]// Biosurfactants. Berlin, Heidelberg: Springer, 2011: 57-91. |
| [5] | Théatre A, Hoste ACR, Rigolet A, et al. Bacillus sp.: a remarkable source of bioactive lipopeptides[M]//Biosurfactants for the Biobased Economy. Cham: Springer International Publishing, 2021: 123-179. |
| [6] |
Arima K, Kakinuma A, Tamura G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation[J]. Biochem Biophys Res Commun, 1968, 31(3): 488-494.
doi: 10.1016/0006-291X(68)90503-2 URL |
| [7] |
Kakinuma A, Hori M, Isono M, et al. Determination of amino acid sequence in surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis[J]. Agric Biol Chem, 1969, 33(6): 971-972.
doi: 10.1080/00021369.1969.10859408 URL |
| [8] |
Kakinuma A, Sugino H, Isono M, et al. Determination of fatty acid in surfactin and elucidation of the total structure of surfactin[J]. Agric Biol Chem, 1969, 33(6): 973-976.
doi: 10.1080/00021369.1969.10859409 URL |
| [9] |
Maget-Dana R, Ptak M. Interfacial properties of surfactin[J]. J Colloid Interface Sci, 1992, 153(1): 285-291.
doi: 10.1016/0021-9797(92)90319-H URL |
| [10] |
Liu JF, Mbadinga SM, Yang SZ, et al. Chemical structure, property and potential applications of biosurfactants produced by Bacillus subtilis in petroleum recovery and spill mitigation[J]. Int J Mol Sci, 2015, 16(3): 4814-4837.
doi: 10.3390/ijms16034814 URL |
| [11] |
Augustyn AR, Pott RWM, Tadie M. The interactions of the biosurfactant surfactin in coal flotation[J]. Colloids Surf A, 2021, 627: 127122.
doi: 10.1016/j.colsurfa.2021.127122 URL |
| [12] |
Deleu M, Bouffioux O, Razafindralambo H, et al. Interaction of surfactin with membranes: a computational approach[J]. Langmuir, 2003, 19(8): 3377-3385.
doi: 10.1021/la026543z URL |
| [13] |
Grau A, Gómez Fernández JC, Peypoux F, et al. A study on the interactions of surfactin with phospholipid vesicles[J]. Biochim Biophys Acta, 1999, 1418(2): 307-319.
pmid: 10320682 |
| [14] |
Chen XY, Zhao HY, Meng FQ, et al. Ameliorated effects of a lipopeptide surfactin on insulin resistance in vitro and in vivo[J]. Food Sci Nutr, 2022, 10(7): 2455-2469.
doi: 10.1002/fsn3.v10.7 URL |
| [15] |
Kotoky R, Pandey P. Rhizosphere mediated biodegradation of benzo(a)pyrene by surfactin producing soil bacilli applied through Melia azadirachta rhizosphere[J]. Int J Phytoremediation, 2020, 22(4): 363-372.
doi: 10.1080/15226514.2019.1663486 URL |
| [16] |
Bais HP, Fall R, Vivanco JM. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production[J]. Plant Physiol, 2004, 134(1): 307-319.
doi: 10.1104/pp.103.028712 URL |
| [17] |
Zhao F, Zhu HB, Cui QF, et al. Anaerobic production of surfactin by a new Bacillus subtilis isolate and the in situ emulsification and viscosity reduction effect towards enhanced oil recovery applications[J]. J Petrol Sci Eng, 2021, 201: 108508.
doi: 10.1016/j.petrol.2021.108508 URL |
| [18] |
Tank JG, Pandya RV. Anti-proliferative activity of surfactins on human cancer cells and their potential use in therapeutics[J]. Peptides, 2022, 155: 170836.
doi: 10.1016/j.peptides.2022.170836 URL |
| [19] |
Meena KR, Sharma A, Kanwar SS. Antitumoral and antimicrobial activity of surfactin extracted from Bacillus subtilis KLP2015[J]. Int J Pept Res Ther, 2020, 26(1): 423-433.
doi: 10.1007/s10989-019-09848-w |
| [20] |
Lim JH, Park BK, Kim MS, et al. The anti-thrombotic activity of surfactins[J]. J Vet Sci, 2005, 6(4): 353-355.
doi: 10.4142/jvs.2005.6.4.353 URL |
| [21] |
Wang Y, Tian JH, Shi FF, et al. Protective effect of surfactin on copper sulfate-induced inflammation, oxidative stress, and hepatic injury in zebrafish[J]. Microbiol Immunol, 2021, 65(10): 410-421.
doi: 10.1111/mim.v65.10 URL |
| [22] |
Mnif I, Segovia R, Bouallegue A, et al. Identification of different lipopeptides isoforms produced by Bacillus mojavensis BI2 and evaluation of their surface activities for potential environmental application[J]. J Polym Environ, 2023, 31(6): 2668-2685.
doi: 10.1007/s10924-022-02752-3 |
| [23] |
Ishikawa F, Konno S, Uchida C, et al. Chemoproteomics profiling of surfactin-producing nonribosomal peptide synthetases in living bacterial cells[J]. Cell Chem Biol, 2022, 29(1): 145-156.e8.
doi: 10.1016/j.chembiol.2021.05.014 URL |
| [24] |
Fortinez CM, Bloudoff K, Harrigan C, et al. Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module[J]. Nat Commun, 2022, 13(1): 548.
doi: 10.1038/s41467-022-28221-y pmid: 35087027 |
| [25] |
Finking R, Mofid MR, Marahiel MA. Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in protein-protein recognition[J]. Biochemistry, 2004, 43(28): 8946-8956.
pmid: 15248752 |
| [26] |
Peypoux F, Bonmatin JM, Labbé H, et al. Isolation and characterization of a new variant of surfactin, the[Val7]surfactin[J]. Eur J Biochem, 1991, 202(1): 101-106.
pmid: 1935967 |
| [27] |
Dowling DP, Kung Y, Croft AK, et al. Structural elements of an NRPS cyclization domain and its inter module docking domain[J]. Proc Natl Acad Sci USA, 2016, 113(44): 12432-12437.
pmid: 27791103 |
| [28] |
Schneider A, Marahiel MA. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis[J]. Arch Microbiol, 1998, 169(5): 404-410.
pmid: 9560421 |
| [29] |
Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics[J]. Chem Rev, 2005, 105(2): 715-738.
pmid: 15700962 |
| [30] |
Tseng CC, Bruner SD, Kohli RM, et al. Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase[J]. Biochemistry, 2002, 41(45): 13350-13359.
pmid: 12416979 |
| [31] |
Bruner SD, Weber T, Kohli RM, et al. Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE[J]. Structure, 2002, 10(3): 301-310.
pmid: 12005429 |
| [32] |
Linne U, Schwarzer D, Schroeder GN, et al. Mutational analysis of a type II thioesterase associated with nonribosomal peptide synthesis[J]. Eur J Biochem, 2004, 271(8): 1536-1545.
pmid: 15066179 |
| [33] |
Schwarzer D, Mootz HD, Linne U, et al. Regeneration of misprimednonribosomal peptide synthetases by type II thioesterases[J]. PNAS, 2002, 99(22): 14083-14088.
pmid: 12384573 |
| [34] |
Chiocchini C, Linne U, Stachelhaus T. In vivo biocombinatorial synthesis of lipopeptides by COM domain-mediated reprogramming of the surfactin biosynthetic complex[J]. Chem Biol, 2006, 13(8): 899-908.
doi: 10.1016/j.chembiol.2006.06.015 URL |
| [35] |
Nakano MM, Zuber P. The primary role of comA in establishment of the competent state in Bacillus subtilis is to activate expression of srfA[J]. J Bacteriol, 1991, 173(22): 7269-7274.
pmid: 1938921 |
| [36] |
Nakano MM, Xia LA, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis[J]. J Bacteriol, 1991, 173(17): 5487-5493.
pmid: 1715856 |
| [37] |
Ogura M, Fujita Y. Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression[J]. FEMS Microbiol Lett, 2007, 268(1): 73-80.
doi: 10.1111/fml.2007.268.issue-1 URL |
| [38] |
Liang Z, Qiao JQ, Li PP, et al. A novel Rap-Phr system in Bacillus velezensis NAU-B3 regulates surfactin production and sporulation via interaction with ComA[J]. Appl Microbiol Biotechnol, 2020, 104(23): 10059-10074.
doi: 10.1007/s00253-020-10942-z pmid: 33043389 |
| [39] |
Nakano MM, Corbell N, Besson J, et al. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis[J]. Mol Gen Genet, 1992, 232(2): 313-321.
doi: 10.1007/BF00280011 |
| [40] |
Tsuge K, Ohata Y, Shoda M. Gene yerP, involved in surfactin self-resistance in Bacillus subtilis[J]. Antimicrob Agents Chemother, 2001, 45(12): 3566-3573.
pmid: 11709341 |
| [41] |
Li X, Yang H, Zhang DL, et al. Overexpression of specific proton motive force-dependent transporters facilitate the export of surfactin in Bacillus subtilis[J]. J Ind Microbiol Biotechnol, 2015, 42(1): 93-103.
doi: 10.1007/s10295-014-1527-z URL |
| [42] | 王镜岩. 生物化学[M]. 第3版. 北京: 高等教育出版社, 2002. |
| Wang JY. Biochemistry[M]. 3rd ed. Beijing: Higher Education Press, 2002. | |
| [43] |
Youssef NH, Wofford N, McInerney MJ. Importance of the long-chain fatty acid beta-hydroxylating cytochrome P450 enzyme YbdT for lipopeptide biosynthesis in Bacillus subtilis strain OKB105[J]. Int J Mol Sci, 2011, 12(3): 1767-1786.
doi: 10.3390/ijms12031767 pmid: 21673922 |
| [44] |
Hlavica P, Lehnerer M. Oxidative biotransformation of fatty acids by cytochromes P450: predicted key structural elements orchestrating substrate specificity, regioselectivity and catalytic efficiency[J]. Curr Drug Metab, 2010, 11(1): 85-104.
pmid: 20302567 |
| [45] |
Steller S, Sokoll A, Wilde C, et al. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein[J]. Biochemistry, 2004, 43(35): 11331-11343.
pmid: 15366943 |
| [46] | Dae KS, Cho J, Park HJ, et al. A comparison of the anti-inflammatory activity of surfactin A, B, C, and D from Bacillus subtilis[J]. J Microbiol Biotechnol, 2006, 16: 1656-1659. |
| [47] |
Bonmatin JM, Labbé H, Grangemard I, et al. Production, isolation and characterization of[Leu4]- and[Ile4]surfactins from Bacillus subtilis[J]. Lett Pept Sci, 1995, 2(1): 41-47.
doi: 10.1007/BF00122922 URL |
| [48] |
Park G, Nam J, Kim J, et al. Structure and mechanism of surfactin peptide from Bacillus velezensis antagonistic to fungi plant pathogens[J]. Bull Korean Chem Soc, 2019, 40(7): 704-709.
doi: 10.1002/bkcs.2019.40.issue-7 URL |
| [49] |
Vater J, Stein T, Vollenbroich D, et al. The modular organization of multifunctional peptide synthetases[J]. J Protein Chem, 1997, 16(5): 557-564.
doi: 10.1023/a:1026386100259 pmid: 9246644 |
| [50] |
Grady EN, MacDonald J, Ho MT, et al. Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6[J]. BMC Microbiol, 2019, 19(1): 5.
doi: 10.1186/s12866-018-1380-8 |
| [51] |
Nerurkar AS. Structural and molecular characteristics of lichenysin and its relationship with surface activity[J]. Adv Exp Med Biol, 2010, 672: 304-315.
pmid: 20545292 |
| [52] |
Naruse N, Tenmyo O, Kobaru S, et al. Pumilacidin, a complex of new antiviral antibiotics. Production, isolation, chemical properties, structure and biological activity[J]. J Antibiot, 1990, 43(3): 267-280.
pmid: 2157695 |
| [53] |
Kecskeméti A, Bartal A, Bóka B, et al. High-frequency occurrence of surfactin monomethyl isoforms in the ferment broth of a Bacillus subtilis strain revealed by ion trap mass spectrometry[J]. Molecules, 2018, 23(9): 2224.
doi: 10.3390/molecules23092224 URL |
| [54] |
Théatre A, Cano-Prieto C, Bartolini M, et al. The surfactin-like lipopeptides from Bacillus spp.: natural biodiversity and synthetic biology for a broader application range[J]. Front Bioeng Biotechnol, 2021, 9: 623701.
doi: 10.3389/fbioe.2021.623701 URL |
| [55] |
Liu XY, Yang SZ, Mu BZ. Production and characterization of a C15-surfactin-O-methyl ester by a lipopeptide producing strain Bacillus subtilis HSO121[J]. Process Biochem, 2009, 44(10): 1144-1151.
doi: 10.1016/j.procbio.2009.06.014 URL |
| [56] |
Li YM, Yang SZ, Mu BZ. Structural characterization of lipopeptide methyl esters produced by Bacillus licheniformis HSN 221[J]. Chem Biodivers, 2010, 7(8): 2065-2075.
doi: 10.1002/cbdv.v7:8 URL |
| [57] | 刘皓, 杨欢, 李雪, 等. 脂肽-糖脂混合生物表面活性剂产生菌筛选和优化培养[J]. 生物工程学报, 2013, 29(12): 1870-1874. |
| Liu H, Yang H, Li X, et al. Identification of Bacillus subtilis THY-7 and high titer optimization for the blend-biosurfactant of lipopeptide and glycolipid[J]. Chin J Biotechnol, 2013, 29(12): 1870-1874.. | |
| [58] |
Wang CY, Cao YX, Wang YP, et al. Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis[J]. Microb Cell Fact, 2019, 18(1): 90.
doi: 10.1186/s12934-019-1139-4 |
| [59] | 周泽宇, 张婉茹, 张柔萱, 等. 代谢工程改造Bacillus amyloliquefaciens提高Surfactin产量[J]. 南开大学学报: 自然科学版, 2018, 51(5): 18-26. |
| Zhou ZY, Zhang WR, Zhang RX, et al. Metabolic engineering of Bacillus amyloliquefaciens to improve surfactin production[J]. Acta Sci Nat Univ Nankaiensis, 2018, 51(5): 18-26. | |
| [60] |
Klausmann P, Hennemann K, Hoffmann M, et al. Bacillus subtilis high cell density fermentation using a sporulation-deficient strain for the production of surfactin[J]. Appl Microbiol Biotechnol, 2021, 105(10): 4141-4151.
doi: 10.1007/s00253-021-11330-x pmid: 33991199 |
| [61] |
Hu FX, Cai WJ, Lin JZ, et al. Genetic engineering of the precursor supply pathway for the overproduction of the nC14-surfactin isoform with promising MEOR applications[J]. Microb Cell Fact, 2021, 20(1): 96.
doi: 10.1186/s12934-021-01585-4 |
| [62] |
Jiang J, Gao L, Bie XM, et al. Identification of novel surfactin derivatives from NRPS modification of Bacillus subtilis and its antifungal activity against Fusarium moniliforme[J]. BMC Microbiol, 2016, 16: 31.
doi: 10.1186/s12866-016-0645-3 pmid: 26957318 |
| [63] |
Eppelmann K, Stachelhaus T, Marahiel MA. Exploitation of the selectivity-conferring code of nonribosomal peptide synthetases for the rational design of novel peptide antibiotics[J]. Biochemistry, 2002, 41(30): 9718-9726.
pmid: 12135394 |
| [64] |
Mootz HD, Kessler N, Linne U, et al. Decreasing the ring size of a cyclic nonribosomal peptide antibiotic by in-frame module deletion in the biosynthetic genes[J]. J Am Chem Soc, 2002, 124(37): 10980-10981.
pmid: 12224936 |
| [65] | 高圣风, 王锋, 刘爱勤, 等. Bacillus subtilis VD18R19脂肽类产物鉴定及其对胡椒花叶病的田间生防效果[J]. 热带农业科学, 2019, 39(10): 89-94. |
| Gao SF, Wang F, Liu AQ, et al. Identification and field application of the lipopeptides produced by Bacillus subtilis VD18R19 against black pepper mosaic disease[J]. Chin J Trop Agric, 2019, 39(10): 89-94. | |
| [66] |
高毓晗, 李世东, 郭荣君. sfp基因转化增强了Bacillus subtilis 168的定殖能力和对黄瓜茎内枯萎病菌的抑制作用[J]. 中国生物防治学报, 2016, 32(1): 76-85.
doi: 10.16409/j.cnki.2095-039x.2015.06.012 |
| Gao YH, Li SD, Guo RJ. Transformation of sfp gene into Bacillus subtilis 168 promotes its colonization on cucumber roots and suppression of Fusarium oxysporum f. sp. cucumerinum in cucumber stems[J]. Chin J Biol Contr, 2016, 32(1): 76-85. | |
| [67] | 周维, 田丹丹, 杨扬, 等. 解淀粉芽孢杆菌G9R-3脂肽类化合物抑制香蕉枯萎病菌机理及防效评价[J]. 西南农业学报, 2019, 32(8): 1810-1816. |
| Zhou W, Tian DD, Yang Y, et al. Antifungal mechanism and control effects on Fusarium oxysporum f. sp. cubense race 4 of lipopeptides produced by Bacillus amyloliquefaciens G9R-3[J]. Southwest China J Agric Sci, 2019, 32(8): 1810-1816. | |
| [68] |
Sarwar A, Hassan MN, Imran M, et al. Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease[J]. Microbiol Res, 2018, 209: 1-13.
doi: 10.1016/j.micres.2018.01.006 URL |
| [69] | Laird M, Piccoli D, Weselowski B, et al. Surfactin-producing Bacillus velezensis 1B-23 and Bacillus sp. 1D-12 protect tomato against bacterial canker caused by Clavibactermichiganensis subsp. michiganensis[J]. J Plant Pathol, 2020, 102(2): 451-458. |
| [70] |
Fan HY, Zhang ZW, Li Y, et al. Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization[J]. Front Microbiol, 2017, 8: 1973.
doi: 10.3389/fmicb.2017.01973 URL |
| [71] |
Li Y, Héloir MC, Zhang X, et al. Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation[J]. Mol Plant Pathol, 2019, 20(8): 1037-1050.
doi: 10.1111/mpp.2019.20.issue-8 URL |
| [72] | 任鹏举, 谢永丽, 张岩, 等. 枯草芽孢杆菌OKB105产生的surfactin防治烟草花叶病毒病及其机理研究[J]. 中国生物防治学报, 2014, 30(2): 216-221. |
| Ren PJ, Xie YL, Zhang Y, et al. Effect and mechanism of controlling TMV disease on tobacco by surfactin produced by Bacillus subtilis OKB105[J]. Chin J Biol Contr, 2014, 30(2): 216-221. | |
| [73] |
Ali SAM, Sayyed RZ, Mir MI, et al. Induction of systemic resistance in maize and antibiofilm activity of surfactin from Bacillus velezensis MS20[J]. Front Microbiol, 2022, 13: 879739.
doi: 10.3389/fmicb.2022.879739 URL |
| [74] |
Le Mire G, Siah A, Brisset MN, et al. Surfactin protects wheat against Zymoseptoriatritici and activates both salicylic acid- and jasmonic acid-dependent defense responses[J]. Agriculture, 2018, 8(1): 11.
doi: 10.3390/agriculture8010011 URL |
| [75] |
Rodríguez J, Tonelli ML, Figueredo MS, et al. The lipopeptide surfactin triggers induced systemic resistance and priming state responses in Arachis hypogaea L.[J]. Eur J Plant Pathol, 2018, 152(3): 845-851.
doi: 10.1007/s10658-018-1524-6 |
| [76] |
Cawoy H, Mariutto M, Henry G, et al. Plant defense stimulation by natural isolates of bacillus depends on efficient surfactin production[J]. Mol Plant Microbe Interact, 2014, 27(2): 87-100.
doi: 10.1094/MPMI-09-13-0262-R URL |
| [77] |
Krishnan N, Velramar B, Velu RK. Investigation of antifungal activity of surfactin against mycotoxigenic phytopathogenic fungus Fusariummoniliforme and its impact in seed germination and mycotoxicosis[J]. Pestic Biochem Physiol, 2019, 155: 101-107.
doi: 10.1016/j.pestbp.2019.01.010 URL |
| [78] |
Wang YY, Zhang CY, Liang J, et al. Surfactin and fengycin B extracted from Bacillus pumilus W-7 provide protection against potato late blight via distinct and synergistic mechanisms[J]. Appl Microbiol Biotechnol, 2020, 104(17): 7467-7481.
doi: 10.1007/s00253-020-10773-y |
| [79] |
Henry G, Deleu M, Jourdan E, et al. The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses[J]. Cell Microbiol, 2011, 13(11): 1824-1837.
doi: 10.1111/j.1462-5822.2011.01664.x pmid: 21838773 |
| [80] | Carrillo C, Teruel JA, Aranda FJ, et al. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin[J]. Biochim Biophys Acta, 2003, 1611(1/2): 91-97. |
| [81] |
Heerklotz H, Seelig J. Leakage and lysis of lipid membranes induced by the lipopeptide surfactin[J]. Eur Biophys J, 2007, 36(4/5): 305-314.
doi: 10.1007/s00249-006-0091-5 URL |
| [82] |
Deravel J, Lemière S, Coutte F, et al. Mycosubtilin and surfactin are efficient, low ecotoxicity molecules for the biocontrol of lettuce downy mildew[J]. Appl Microbiol Biotechnol, 2014, 98(14): 6255-6264.
doi: 10.1007/s00253-014-5663-1 pmid: 24723290 |
| [83] | de Andrade CJ, Barros FF, de Andrade LM, et al. Ultrafiltration based purification strategies for surfactin produced by Bacillus subtilis LB5A using cassava wastewater as substrate[J]. J Chem Technol Biotechnol, 2016, 91(12): 3018-3027. |
| [84] |
Marin CP, Kaschuk JJ, Frollini E, et al. Potential use of the liquor from sisal pulp hydrolysis as substrate for surfactin production[J]. Ind Crops Prod, 2015, 66: 239-245.
doi: 10.1016/j.indcrop.2015.01.001 URL |
| [85] |
Zanotto AW, Valério A, de Andrade CJ, et al. New sustainable alternatives to reduce the production costs for surfactin 50 years after the discovery[J]. Appl Microbiol Biotechnol, 2019, 103(21/22): 8647-8656.
doi: 10.1007/s00253-019-10123-7 |
| [86] |
de Oliveira Schmidt VK, Vicente R, et al. Cassava wastewater valorization for the production of biosurfactants: surfactin, rhamnolipids, and mannosileritritol lipids[J]. World J Microbiol Biotechnol, 2022, 39(2): 65.
doi: 10.1007/s11274-022-03510-2 |
| [87] |
Kisil OV, Trefilov VS, Sadykova VS, et al. Surfactin: its biological activity and possibility of application in agriculture[J]. Appl Biochem Microbiol, 2023, 59(1): 1-13.
doi: 10.1134/S0003683823010027 |
| [1] | 王子璇, 隋玉, 尚学钰, 马思嘉, 吴天香, 刘洋, 王琦. 元蘑多糖的结构特征及其调节巨噬细胞免疫活性机制[J]. 生物技术通报, 2024, 40(1): 308-321. |
| [2] | 陈治民, 李翠, 韦继天, 李昕然, 刘峄, 郭强. 绿原酸生物合成调控及其应用研究进展[J]. 生物技术通报, 2024, 40(1): 57-71. |
| [3] | 李亮, 徐姗姗, 姜艳军. 生物合成法生产麦角硫因的研究进展[J]. 生物技术通报, 2024, 40(1): 86-99. |
| [4] | 褚睿, 李昭轩, 张学青, 杨东亚, 曹行行, 张雪艳. 黄瓜枯萎病拮抗芽孢杆菌的筛选、鉴定及其生防潜力[J]. 生物技术通报, 2023, 39(8): 262-271. |
| [5] | 叶云芳, 田清尹, 施婷婷, 王亮, 岳远征, 杨秀莲, 王良桂. 植物中β-紫罗兰酮生物合成及调控研究进展[J]. 生物技术通报, 2023, 39(8): 91-105. |
| [6] | 张曼, 张叶卓, 何其邹洪, 鄂一岚, 李晔. 植物细胞壁结构及成像技术研究进展[J]. 生物技术通报, 2023, 39(7): 113-122. |
| [7] | 王玲, 卓燊, 付学森, 刘紫璇, 刘笑蓉, 王志辉, 周日宝, 刘湘丹. 莲生物碱生物合成途径及相关基因研究进展[J]. 生物技术通报, 2023, 39(7): 56-66. |
| [8] | 崔学强, 黄昌艳, 邓杰玲, 李先民, 李秀玲, 张自斌. 基于SLAF-seq技术的石斛兰SNP标记开发及亲缘关系分析[J]. 生物技术通报, 2023, 39(6): 141-148. |
| [9] | 李托, 李陇平, 屈雷. 有尾噬菌体的结构及其受体研究进展[J]. 生物技术通报, 2023, 39(6): 88-101. |
| [10] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [11] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [12] | 周定定, 李辉虎, 汤兴涌, 余发新, 孔丹宇, 刘毅. 甘草酸和甘草苷生物合成与调控的研究进展[J]. 生物技术通报, 2023, 39(5): 44-53. |
| [13] | 郁慧丽, 李爱涛. 细胞色素P450酶在香精香料绿色生物合成中的应用[J]. 生物技术通报, 2023, 39(4): 24-37. |
| [14] | 章乐乐, 王冠, 柳凤, 胡汉桥, 任磊. 芒果炭疽病拮抗菌分离、鉴定及生防机制研究[J]. 生物技术通报, 2023, 39(4): 277-287. |
| [15] | 杨俊钊, 张新蕊, 赵国柱, 郑菲. 新型GH5家族多结构域纤维素酶的结构与功能研究[J]. 生物技术通报, 2023, 39(4): 71-80. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||