








生物技术通报 ›› 2024, Vol. 40 ›› Issue (11): 142-151.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0297
李晴1( ), 石雨荷1, 朱珏1, 李晓玲1, 侯超文1, 童巧珍1,2,3(
), 石雨荷1, 朱珏1, 李晓玲1, 侯超文1, 童巧珍1,2,3( )
)
收稿日期:2024-03-26
出版日期:2024-11-26
发布日期:2024-12-19
通讯作者:
童巧珍,女,博士,教授,博士生导师,研究方向:中药资源与品质评价;E-mail: qztong88@126.com作者简介:李晴,女,硕士研究生,研究方向:中药资源与开发;E-mail: 984912094@qq.com
基金资助:
LI Qing1( ), SHI Yu-he1, ZHU Jue1, LI Xiao-ling1, HOU Chao-wen1, TONG Qiao-zhen1,2,3(
), SHI Yu-he1, ZHU Jue1, LI Xiao-ling1, HOU Chao-wen1, TONG Qiao-zhen1,2,3( )
)
Received:2024-03-26
Published:2024-11-26
Online:2024-12-19
摘要:
【目的】 采用SCoT分子标记技术对17份白术种质资源进行遗传多样性分析及DNA指纹图谱构建,为白术种质的鉴定、保存和新品种选育提供一定的理论依据。【方法】 从92条SCoT引物中筛选得到13条核心引物用于白术分子标记,利用Popgene1.32软件和NTSYS-pc 2.10e软件进行白术种质资源多样性分析和聚类分析,以核心引物SCoT-9、SCoT-12和SCoT-41组合构建白术DNA分子指纹图谱。【结果】 13条SCoT核心引物扩增17份白术样品共得到192条条带,其中,多态性条带165条,平均百分率85.94%;遗传相似系数(GS)和遗传距离(GD)分别介于0.578 1-0.875 0和0.133 5-0.548 0,白术种质的观测等位基因数(Na)为1.859 4,有效等位基因数(Ne)为1.418 0,Nei's遗传多样性指数(He)为0.256 3,Shannon信息指数(I)为0.396 8,聚类分析得到白术栽培品一类,而大围山野生品被单独聚为一类;所构建的17份白术种质资源DNA指纹图谱,可以将样品区分并准确鉴定。【结论】 白术种质资源具有较为丰富的遗传多样性,但不同产地白术栽培品之间遗传差异低,表明地理位置并不是判断白术亲缘关系远近的决定性因素。
李晴, 石雨荷, 朱珏, 李晓玲, 侯超文, 童巧珍. 基于SCoT分子标记分析白术种质资源遗传多样性及DNA指纹图谱构建[J]. 生物技术通报, 2024, 40(11): 142-151.
LI Qing, SHI Yu-he, ZHU Jue, LI Xiao-ling, HOU Chao-wen, TONG Qiao-zhen. Genetic Diversity Analysis and DNA Fingerprint Construction of Atractylodes macrocephala Germplasm Resources Based on SCoT Molecular Markers[J]. Biotechnology Bulletin, 2024, 40(11): 142-151.
| 编号 ID | 种源地 Provenance | 类型 Type | 采收地 Field of harvest | 采收时间 Harvest time |
|---|---|---|---|---|
| BZ-1 | 安徽亳州1号 Bozhou No.1, Anhui | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-2 | 安徽亳州2号 Bozhou No.2, Anhui | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-3 | 河北安国1号 Anguo No.1, Hebei | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-4 | 河北安国2号 Anguo No.2, Hebei | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-5 | 湖南平江龙门镇/新和村 Pingjiang Longmen Town, Hunan/Xinhe Village | 栽培 Cultivating | 药植园资源圃Medicinal plantation resource garden | 2021-04 |
| BZ-6 | 湖南平江龙门镇/泉水村 Pingjiang Longmen Town, Hunan/Quanshui Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-7 | 湖南平江龙门镇/丰福村 Pingjiang Longmen Town, Hunan/Fengfu Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-8 | 湖南平江龙门镇/福寿村 Pingjiang Longmen Town, Hunan/Fushou Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-9 | 湖南平江龙门镇/渣坪村 Pingjiang Longmen Town, Hunan/Zhaping Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-10 | 湖南衡阳祁东 Hengyang Qidong, Hunan | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-11 | 湖南岳阳临湘 Yueyang Linxiang, Hunan | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-12 | 湖南怀化 Huaihua, Hunan | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-13 | 江西修水 Xiushui, Jiangxi | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-14 | 湖北恩施 Enshi, Hubei | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-15 | 浙江于潜1号 Yuqian No.1, Zhejiang | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-16 | 浙江于潜2号 Yuqian No.2, Zhejiang | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-17 | 湖南浏阳大围山 Liuyang Dawei Mountain, Hunan | 野生 Wild | 药植园资源圃 Medicinal plantation resource garden | 2021-06 |
表1 供试白术材料及其来源
Table 1 Test materials and sources of A. macrocephala
| 编号 ID | 种源地 Provenance | 类型 Type | 采收地 Field of harvest | 采收时间 Harvest time |
|---|---|---|---|---|
| BZ-1 | 安徽亳州1号 Bozhou No.1, Anhui | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-2 | 安徽亳州2号 Bozhou No.2, Anhui | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-3 | 河北安国1号 Anguo No.1, Hebei | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-4 | 河北安国2号 Anguo No.2, Hebei | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-5 | 湖南平江龙门镇/新和村 Pingjiang Longmen Town, Hunan/Xinhe Village | 栽培 Cultivating | 药植园资源圃Medicinal plantation resource garden | 2021-04 |
| BZ-6 | 湖南平江龙门镇/泉水村 Pingjiang Longmen Town, Hunan/Quanshui Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-7 | 湖南平江龙门镇/丰福村 Pingjiang Longmen Town, Hunan/Fengfu Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-8 | 湖南平江龙门镇/福寿村 Pingjiang Longmen Town, Hunan/Fushou Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-9 | 湖南平江龙门镇/渣坪村 Pingjiang Longmen Town, Hunan/Zhaping Village | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-10 | 湖南衡阳祁东 Hengyang Qidong, Hunan | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-11 | 湖南岳阳临湘 Yueyang Linxiang, Hunan | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-12 | 湖南怀化 Huaihua, Hunan | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-13 | 江西修水 Xiushui, Jiangxi | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-14 | 湖北恩施 Enshi, Hubei | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-15 | 浙江于潜1号 Yuqian No.1, Zhejiang | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-16 | 浙江于潜2号 Yuqian No.2, Zhejiang | 栽培 Cultivating | 药植园资源圃 Medicinal plantation resource garden | 2021-04 |
| BZ-17 | 湖南浏阳大围山 Liuyang Dawei Mountain, Hunan | 野生 Wild | 药植园资源圃 Medicinal plantation resource garden | 2021-06 |
| 引物编码Primer name | 序列Sequence(5'-3') | 引物编码Primer name | 序列Sequence(5'-3') | |
|---|---|---|---|---|
| SCoT-1 | ACCATGGCTACCACCGCG | SCoT-47 | CCATGGCTACCACCGGCA | |
| SCoT-2 | ACGACATGGCGACCGCGA | SCoT-48 | CAGCAATGGCTACCACCA | |
| SCoT-3 | AAGCAATGGCTACCACCA | SCoT-49 | TCGACATGGCGACCCACG | |
| SCoT-4 | ACGACATGGCGACCATCG | SCoT-50 | GAACAATGGCTACCACGC | |
| SCoT-5 | ACGACATGGCGACCACGC | SCoT-51 | GCAACAATGGCTACCACC | |
| SCoT-6 | ACCATGGCTACCACCGGC | SCoT-52 | GCAACAATGGCTACCACG | |
| SCoT-7 | ACCATGGCTACCACCGCA | SCoT-53 | AACCATGGCTACCACCAC | |
| SCoT-8 | ACGACATGGCGACCAACG | SCoT-54 | AAGCAATGCGTACCACCA | |
| SCoT-9 | ACGACATGGCGACCACGT | SCoT-55 | ACAATGGCTACCACCACA | |
| SCoT-10 | ACAATGGCTACCACTGCC | SCoT-56 | ACAATGGCTACCACCAGC | |
| SCoT-11 | ACCATGGCTACCAGCGGC | SCoT-57 | ACAATGGCTACCACCATC | |
| SCoT-12 | ACCATGGCTACCAGCGCA | SCoT-58 | ACAATGGCTACCACCGAC | |
| SCoT-13 | ACCATGGCTACCAGCGCG | SCoT-59 | ACAATGGCTACCACTACC | |
| SCoT-14 | ACGACATGGCGACCAGCG | SCoT-60 | ACAATGGCTACCACTACG | |
| SCoT-15 | ACCATGGCTACCACCGGG | SCoT-61 | ACAATGGCTACCACTAGC | |
| SCoT-16 | ACCATGGCTACCACCGTC | SCoT-62 | ACAATGGCTACCACTGAC | |
| SCoT-17 | ACCATGGCTACCACCGTG | SCoT-63 | ACAATGGCTACCACTGAG | |
| SCoT-18 | AAGCAATGGCTACCACCG | SCoT-64 | ACAATGGCTACCACTGCA | |
| SCoT-19 | ACCATGGCTACCACCGAC | SCoT-65 | ACAATGGCTACCACTGGC | |
| SCoT-20 | ACCATGGCTACCACCGAG | SCoT-66 | ACAATGGCTACCACTGGG | |
| SCoT-21 | ACCATGGCTACCACCGCC | SCoT-67 | ACAATGGCTACCACTGTC | |
| SCoT-22 | ACGACATGGCGACCGCGG | SCoT-68 | ACCATGGCTACCACCGCT | |
| SCoT-23 | ACCATGGCTACCACCGAT | SCoT-69 | ACCATGGCTACCACGGAG | |
| SCoT-24 | ACGACATGGCGACCCACA | SCoT-70 | ACCATGGCTACCACGGCA | |
| SCoT-25 | AACCATGGTCACCACCAC | SCoT-71 | ACCATGGCTACCACGGGC | |
| SCoT-26 | ACGACATGGCGACCATCT | SCoT-72 | ACCATGGCTACCACGGTC | |
| SCoT-27 | ATGACATGGCGACCATGC | SCoT-73 | ACCATGGCTACCAGCGAG | |
| SCoT-28 | CAACAATGGCTACCACGT | SCoT-74 | ACCATGGCTACCAGCGTC | |
| SCoT-29 | CCATGGCTACCACCGGCC | SCoT-75 | CAACAATGCCTACCACCT | |
| SCoT-30 | CCATGGCTACCACCGGCG | SCoT-76 | CAATGGCTACCACCGCAG | |
| SCoT-31 | CCATGGCTACCACCGCCT | SCoT-77 | CAATGGCTACCACTAACG | |
| SCoT-32 | CCATGGCTACCACCGCAG | SCoT-78 | CAATGGCTACCACTACAG | |
| SCoT-33 | CCATGGCTACCACCGCAC | SCoT-79 | CAATGGCTACCACTAGCC | |
| SCoT-34 | CAACAATGGCTACCACCA | SCoT-80 | CAATGGCTACCACTAGCG | |
| SCoT-35 | CACCATGGCTACCACCAG | SCoT-81 | CAATGGCTACCACTGACA | |
| SCoT-36 | CAACAATGGCTACCACCC | SCoT-82 | CAATGGCTACCATTAGCC | |
| SCoT-37 | CACCATGGCTACCACCAT | SCoT-83 | CAATGGCTACCATTAGCG | |
| SCoT-38 | CAACAATGGCTACCACCG | SCoT-84 | CCATGGCTACCACCGCCC | |
| SCoT-39 | CAACAATGGCTACCACCT | SCoT-85 | CCATGGCTACCACCGCCG | |
| SCoT-40 | CAACAATGGCTACCACGA | SCoT-86 | CCATGGCTACCACCGGAG | |
| SCoT-41 | CAACAATGGCTACCACGC | SCoT-87 | CCATGGCTACCACCGGCT | |
| SCoT-42 | CCATGGCTACCACCGCCA | SCoT-88 | CCATGGCTACCACTACCC | |
| SCoT-43 | CAACAATGGCTACCACGG | SCoT-89 | CCATGGCTACCACTACCG | |
| SCoT-44 | CAACAATGGCTACCAGCA | SCoT-90 | CCATGGCTACCACTAGCG | |
| SCoT-45 | CAACAATGGCTACCAGCC | SCoT-91 | CCATGGCTACCACTAGCT | |
| SCoT-46 | CATGGCTACCACCGGCCC | SCoT-92 | GCCAGCCACCATGGCACA |
表2 SCoT引物的序列
Table 2 Sequences of SCoT primers
| 引物编码Primer name | 序列Sequence(5'-3') | 引物编码Primer name | 序列Sequence(5'-3') | |
|---|---|---|---|---|
| SCoT-1 | ACCATGGCTACCACCGCG | SCoT-47 | CCATGGCTACCACCGGCA | |
| SCoT-2 | ACGACATGGCGACCGCGA | SCoT-48 | CAGCAATGGCTACCACCA | |
| SCoT-3 | AAGCAATGGCTACCACCA | SCoT-49 | TCGACATGGCGACCCACG | |
| SCoT-4 | ACGACATGGCGACCATCG | SCoT-50 | GAACAATGGCTACCACGC | |
| SCoT-5 | ACGACATGGCGACCACGC | SCoT-51 | GCAACAATGGCTACCACC | |
| SCoT-6 | ACCATGGCTACCACCGGC | SCoT-52 | GCAACAATGGCTACCACG | |
| SCoT-7 | ACCATGGCTACCACCGCA | SCoT-53 | AACCATGGCTACCACCAC | |
| SCoT-8 | ACGACATGGCGACCAACG | SCoT-54 | AAGCAATGCGTACCACCA | |
| SCoT-9 | ACGACATGGCGACCACGT | SCoT-55 | ACAATGGCTACCACCACA | |
| SCoT-10 | ACAATGGCTACCACTGCC | SCoT-56 | ACAATGGCTACCACCAGC | |
| SCoT-11 | ACCATGGCTACCAGCGGC | SCoT-57 | ACAATGGCTACCACCATC | |
| SCoT-12 | ACCATGGCTACCAGCGCA | SCoT-58 | ACAATGGCTACCACCGAC | |
| SCoT-13 | ACCATGGCTACCAGCGCG | SCoT-59 | ACAATGGCTACCACTACC | |
| SCoT-14 | ACGACATGGCGACCAGCG | SCoT-60 | ACAATGGCTACCACTACG | |
| SCoT-15 | ACCATGGCTACCACCGGG | SCoT-61 | ACAATGGCTACCACTAGC | |
| SCoT-16 | ACCATGGCTACCACCGTC | SCoT-62 | ACAATGGCTACCACTGAC | |
| SCoT-17 | ACCATGGCTACCACCGTG | SCoT-63 | ACAATGGCTACCACTGAG | |
| SCoT-18 | AAGCAATGGCTACCACCG | SCoT-64 | ACAATGGCTACCACTGCA | |
| SCoT-19 | ACCATGGCTACCACCGAC | SCoT-65 | ACAATGGCTACCACTGGC | |
| SCoT-20 | ACCATGGCTACCACCGAG | SCoT-66 | ACAATGGCTACCACTGGG | |
| SCoT-21 | ACCATGGCTACCACCGCC | SCoT-67 | ACAATGGCTACCACTGTC | |
| SCoT-22 | ACGACATGGCGACCGCGG | SCoT-68 | ACCATGGCTACCACCGCT | |
| SCoT-23 | ACCATGGCTACCACCGAT | SCoT-69 | ACCATGGCTACCACGGAG | |
| SCoT-24 | ACGACATGGCGACCCACA | SCoT-70 | ACCATGGCTACCACGGCA | |
| SCoT-25 | AACCATGGTCACCACCAC | SCoT-71 | ACCATGGCTACCACGGGC | |
| SCoT-26 | ACGACATGGCGACCATCT | SCoT-72 | ACCATGGCTACCACGGTC | |
| SCoT-27 | ATGACATGGCGACCATGC | SCoT-73 | ACCATGGCTACCAGCGAG | |
| SCoT-28 | CAACAATGGCTACCACGT | SCoT-74 | ACCATGGCTACCAGCGTC | |
| SCoT-29 | CCATGGCTACCACCGGCC | SCoT-75 | CAACAATGCCTACCACCT | |
| SCoT-30 | CCATGGCTACCACCGGCG | SCoT-76 | CAATGGCTACCACCGCAG | |
| SCoT-31 | CCATGGCTACCACCGCCT | SCoT-77 | CAATGGCTACCACTAACG | |
| SCoT-32 | CCATGGCTACCACCGCAG | SCoT-78 | CAATGGCTACCACTACAG | |
| SCoT-33 | CCATGGCTACCACCGCAC | SCoT-79 | CAATGGCTACCACTAGCC | |
| SCoT-34 | CAACAATGGCTACCACCA | SCoT-80 | CAATGGCTACCACTAGCG | |
| SCoT-35 | CACCATGGCTACCACCAG | SCoT-81 | CAATGGCTACCACTGACA | |
| SCoT-36 | CAACAATGGCTACCACCC | SCoT-82 | CAATGGCTACCATTAGCC | |
| SCoT-37 | CACCATGGCTACCACCAT | SCoT-83 | CAATGGCTACCATTAGCG | |
| SCoT-38 | CAACAATGGCTACCACCG | SCoT-84 | CCATGGCTACCACCGCCC | |
| SCoT-39 | CAACAATGGCTACCACCT | SCoT-85 | CCATGGCTACCACCGCCG | |
| SCoT-40 | CAACAATGGCTACCACGA | SCoT-86 | CCATGGCTACCACCGGAG | |
| SCoT-41 | CAACAATGGCTACCACGC | SCoT-87 | CCATGGCTACCACCGGCT | |
| SCoT-42 | CCATGGCTACCACCGCCA | SCoT-88 | CCATGGCTACCACTACCC | |
| SCoT-43 | CAACAATGGCTACCACGG | SCoT-89 | CCATGGCTACCACTACCG | |
| SCoT-44 | CAACAATGGCTACCAGCA | SCoT-90 | CCATGGCTACCACTAGCG | |
| SCoT-45 | CAACAATGGCTACCAGCC | SCoT-91 | CCATGGCTACCACTAGCT | |
| SCoT-46 | CATGGCTACCACCGGCCC | SCoT-92 | GCCAGCCACCATGGCACA |
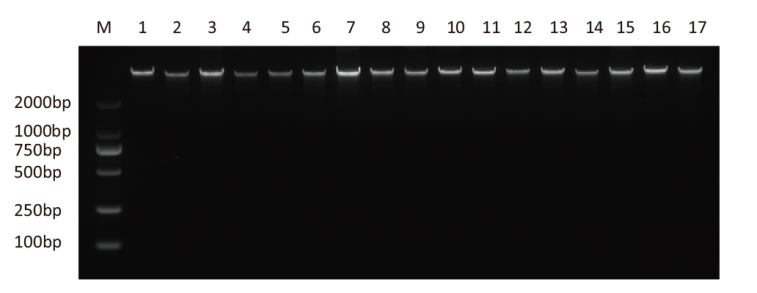
图1 17份白术种质基因组DNA电泳图 M为DL 2000 DNA marker;1-17为17份白术种质编号。下同
Fig. 1 Electrophoresis of genomic DNA of 17 A. macroce-phala germplasms M is the DL 2000 DNA marker; 1-17 are the 17 germplasm numbers of A. macrocephala. The same below
| 引物编号 Primer name | 扩增条带 Total bands | 多态性条带 Polymorphic bands | 多态性比例 Polymorphic percentage/% | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| SCoT-1 | 13 | 11 | 84.62 | 55.8 |
| SCoT-9 | 17 | 14 | 82.35 | 61.3 |
| SCoT-12 | 18 | 17 | 94.44 | 61.3 |
| SCoT-17 | 12 | 11 | 91.67 | 59.9 |
| SCoT-32 | 19 | 17 | 89.47 | 59.9 |
| SCoT-35 | 12 | 11 | 91.67 | 54.4 |
| SCoT-39 | 17 | 12 | 70.59 | 54.4 |
| SCoT-41 | 15 | 14 | 93.33 | 55.8 |
| SCoT-49 | 14 | 11 | 78.57 | 59.9 |
| SCoT-50 | 15 | 14 | 93.33 | 57.2 |
| SCoT-61 | 12 | 11 | 91.67 | 55.8 |
| SCoT-69 | 13 | 11 | 84.62 | 54.6 |
| SCoT-76 | 15 | 11 | 73.33 | 54.6 |
| 总计 Total | 192 | 165 | 85.94 |
表3 13条 SCoT 引物扩增结果
Table 3 Amplification results of 13 SCoT primers
| 引物编号 Primer name | 扩增条带 Total bands | 多态性条带 Polymorphic bands | 多态性比例 Polymorphic percentage/% | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| SCoT-1 | 13 | 11 | 84.62 | 55.8 |
| SCoT-9 | 17 | 14 | 82.35 | 61.3 |
| SCoT-12 | 18 | 17 | 94.44 | 61.3 |
| SCoT-17 | 12 | 11 | 91.67 | 59.9 |
| SCoT-32 | 19 | 17 | 89.47 | 59.9 |
| SCoT-35 | 12 | 11 | 91.67 | 54.4 |
| SCoT-39 | 17 | 12 | 70.59 | 54.4 |
| SCoT-41 | 15 | 14 | 93.33 | 55.8 |
| SCoT-49 | 14 | 11 | 78.57 | 59.9 |
| SCoT-50 | 15 | 14 | 93.33 | 57.2 |
| SCoT-61 | 12 | 11 | 91.67 | 55.8 |
| SCoT-69 | 13 | 11 | 84.62 | 54.6 |
| SCoT-76 | 15 | 11 | 73.33 | 54.6 |
| 总计 Total | 192 | 165 | 85.94 |
| pop ID | BZ-1 | BZ-2 | BZ-3 | BZ-4 | BZ-5 | BZ-6 | BZ-7 | BZ-8 | BZ-9 | BZ-10 | BZ-11 | BZ-12 | BZ-13 | BZ-14 | BZ-15 | BZ-16 | BZ-17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BZ-1 | **** | 0.776 0 | 0.786 5 | 0.750 0 | 0.677 1 | 0.713 5 | 0.729 2 | 0.724 0 | 0.755 2 | 0.697 9 | 0.708 3 | 0.692 7 | 0.692 7 | 0.651 0 | 0.729 2 | 0.692 7 | 0.625 0 |
| BZ-2 | 0.253 5 | **** | 0.812 5 | 0.755 2 | 0.692 7 | 0.739 6 | 0.755 2 | 0.760 4 | 0.708 3 | 0.692 7 | 0.713 5 | 0.697 9 | 0.760 4 | 0.739 6 | 0.786 5 | 0.750 0 | 0.692 7 |
| BZ-3 | 0.240 2 | 0.207 6 | **** | 0.786 5 | 0.744 8 | 0.791 7 | 0.796 9 | 0.781 2 | 0.833 3 | 0.713 5 | 0.765 6 | 0.802 1 | 0.802 1 | 0.760 4 | 0.765 6 | 0.791 7 | 0.661 5 |
| BZ-4 | 0.287 7 | 0.280 8 | 0.240 2 | **** | 0.739 6 | 0.734 4 | 0.760 4 | 0.724 0 | 0.755 2 | 0.677 1 | 0.739 6 | 0.755 2 | 0.713 5 | 0.703 1 | 0.729 2 | 0.744 8 | 0.625 0 |
| BZ-5 | 0.390 0 | 0.367 1 | 0.294 7 | 0.301 7 | **** | 0.724 0 | 0.718 8 | 0.734 4 | 0.734 4 | 0.708 3 | 0.708 3 | 0.692 7 | 0.713 5 | 0.692 7 | 0.750 0 | 0.703 1 | 0.593 8 |
| BZ-6 | 0.337 5 | 0.301 7 | 0.233 6 | 0.308 7 | 0.323 0 | **** | 0.786 5 | 0.760 4 | 0.781 2 | 0.692 7 | 0.744 8 | 0.760 4 | 0.729 2 | 0.697 9 | 0.671 9 | 0.687 5 | 0.588 5 |
| BZ-7 | 0.315 9 | 0.280 8 | 0.227 1 | 0.273 9 | 0.330 2 | 0.240 2 | **** | 0.692 7 | 0.828 1 | 0.697 9 | 0.760 4 | 0.744 8 | 0.786 5 | 0.776 0 | 0.760 4 | 0.734 4 | 0.614 6 |
| BZ-8 | 0.323 0 | 0.273 9 | 0.246 9 | 0.323 0 | 0.308 7 | 0.273 9 | 0.367 1 | **** | 0.760 4 | 0.682 3 | 0.724 0 | 0.739 6 | 0.739 6 | 0.687 5 | 0.724 0 | 0.760 4 | 0.661 5 |
| BZ-9 | 0.280 8 | 0.344 8 | 0.182 3 | 0.280 8 | 0.308 7 | 0.246 9 | 0.188 6 | 0.273 9 | **** | 0.692 7 | 0.765 6 | 0.760 4 | 0.781 2 | 0.729 2 | 0.724 0 | 0.729 2 | 0.578 1 |
| BZ-10 | 0.359 7 | 0.367 1 | 0.337 5 | 0.390 0 | 0.344 8 | 0.367 1 | 0.359 7 | 0.382 3 | 0.367 1 | **** | 0.677 1 | 0.682 3 | 0.640 6 | 0.661 5 | 0.718 8 | 0.661 5 | 0.614 6 |
| BZ-11 | 0.344 8 | 0.337 5 | 0.267 1 | 0.301 7 | 0.344 8 | 0.294 7 | 0.273 9 | 0.323 0 | 0.267 1 | 0.390 0 | **** | 0.838 5 | 0.786 5 | 0.776 0 | 0.729 2 | 0.744 8 | 0.656 2 |
| BZ-12 | 0.367 1 | 0.359 7 | 0.220 5 | 0.280 8 | 0.367 1 | 0.273 9 | 0.294 7 | 0.301 7 | 0.273 9 | 0.382 3 | 0.176 1 | **** | 0.812 5 | 0.791 7 | 0.734 4 | 0.729 2 | 0.630 2 |
| BZ-13 | 0.367 1 | 0.273 9 | 0.220 5 | 0.337 5 | 0.337 5 | 0.315 9 | 0.240 2 | 0.301 7 | 0.246 9 | 0.445 3 | 0.240 2 | 0.207 6 | **** | 0.875 0 | 0.776 0 | 0.760 4 | 0.651 0 |
| BZ-14 | 0.429 2 | 0.301 7 | 0.273 9 | 0.352 2 | 0.367 1 | 0.359 7 | 0.253 5 | 0.374 7 | 0.315 9 | 0.413 3 | 0.253 5 | 0.233 6 | 0.133 5 | **** | 0.786 5 | 0.750 0 | 0.682 3 |
| BZ-15 | 0.315 9 | 0.240 2 | 0.267 1 | 0.315 9 | 0.287 7 | 0.397 7 | 0.273 9 | 0.323 0 | 0.323 0 | 0.330 2 | 0.315 9 | 0.308 7 | 0.253 5 | 0.240 2 | **** | 0.776 0 | 0.677 1 |
| BZ-16 | 0.367 1 | 0.287 7 | 0.233 6 | 0.294 7 | 0.352 2 | 0.374 7 | 0.308 7 | 0.273 9 | 0.315 9 | 0.413 3 | 0.294 7 | 0.315 9 | 0.273 9 | 0.287 7 | 0.253 5 | **** | 0.682 3 |
| BZ-17 | 0.470 0 | 0.367 1 | 0.413 3 | 0.470 0 | 0.521 3 | 0.530 1 | 0.486 8 | 0.413 3 | 0.548 0 | 0.486 8 | 0.421 2 | 0.461 7 | 0.429 2 | 0.382 3 | 0.390 0 | 0.382 3 | **** |
表4 17份白术种质遗传相似系数和遗传距离
Table 4 Genetic similarity coefficients and genetic distances of 17 A. macrocephala germplasms
| pop ID | BZ-1 | BZ-2 | BZ-3 | BZ-4 | BZ-5 | BZ-6 | BZ-7 | BZ-8 | BZ-9 | BZ-10 | BZ-11 | BZ-12 | BZ-13 | BZ-14 | BZ-15 | BZ-16 | BZ-17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BZ-1 | **** | 0.776 0 | 0.786 5 | 0.750 0 | 0.677 1 | 0.713 5 | 0.729 2 | 0.724 0 | 0.755 2 | 0.697 9 | 0.708 3 | 0.692 7 | 0.692 7 | 0.651 0 | 0.729 2 | 0.692 7 | 0.625 0 |
| BZ-2 | 0.253 5 | **** | 0.812 5 | 0.755 2 | 0.692 7 | 0.739 6 | 0.755 2 | 0.760 4 | 0.708 3 | 0.692 7 | 0.713 5 | 0.697 9 | 0.760 4 | 0.739 6 | 0.786 5 | 0.750 0 | 0.692 7 |
| BZ-3 | 0.240 2 | 0.207 6 | **** | 0.786 5 | 0.744 8 | 0.791 7 | 0.796 9 | 0.781 2 | 0.833 3 | 0.713 5 | 0.765 6 | 0.802 1 | 0.802 1 | 0.760 4 | 0.765 6 | 0.791 7 | 0.661 5 |
| BZ-4 | 0.287 7 | 0.280 8 | 0.240 2 | **** | 0.739 6 | 0.734 4 | 0.760 4 | 0.724 0 | 0.755 2 | 0.677 1 | 0.739 6 | 0.755 2 | 0.713 5 | 0.703 1 | 0.729 2 | 0.744 8 | 0.625 0 |
| BZ-5 | 0.390 0 | 0.367 1 | 0.294 7 | 0.301 7 | **** | 0.724 0 | 0.718 8 | 0.734 4 | 0.734 4 | 0.708 3 | 0.708 3 | 0.692 7 | 0.713 5 | 0.692 7 | 0.750 0 | 0.703 1 | 0.593 8 |
| BZ-6 | 0.337 5 | 0.301 7 | 0.233 6 | 0.308 7 | 0.323 0 | **** | 0.786 5 | 0.760 4 | 0.781 2 | 0.692 7 | 0.744 8 | 0.760 4 | 0.729 2 | 0.697 9 | 0.671 9 | 0.687 5 | 0.588 5 |
| BZ-7 | 0.315 9 | 0.280 8 | 0.227 1 | 0.273 9 | 0.330 2 | 0.240 2 | **** | 0.692 7 | 0.828 1 | 0.697 9 | 0.760 4 | 0.744 8 | 0.786 5 | 0.776 0 | 0.760 4 | 0.734 4 | 0.614 6 |
| BZ-8 | 0.323 0 | 0.273 9 | 0.246 9 | 0.323 0 | 0.308 7 | 0.273 9 | 0.367 1 | **** | 0.760 4 | 0.682 3 | 0.724 0 | 0.739 6 | 0.739 6 | 0.687 5 | 0.724 0 | 0.760 4 | 0.661 5 |
| BZ-9 | 0.280 8 | 0.344 8 | 0.182 3 | 0.280 8 | 0.308 7 | 0.246 9 | 0.188 6 | 0.273 9 | **** | 0.692 7 | 0.765 6 | 0.760 4 | 0.781 2 | 0.729 2 | 0.724 0 | 0.729 2 | 0.578 1 |
| BZ-10 | 0.359 7 | 0.367 1 | 0.337 5 | 0.390 0 | 0.344 8 | 0.367 1 | 0.359 7 | 0.382 3 | 0.367 1 | **** | 0.677 1 | 0.682 3 | 0.640 6 | 0.661 5 | 0.718 8 | 0.661 5 | 0.614 6 |
| BZ-11 | 0.344 8 | 0.337 5 | 0.267 1 | 0.301 7 | 0.344 8 | 0.294 7 | 0.273 9 | 0.323 0 | 0.267 1 | 0.390 0 | **** | 0.838 5 | 0.786 5 | 0.776 0 | 0.729 2 | 0.744 8 | 0.656 2 |
| BZ-12 | 0.367 1 | 0.359 7 | 0.220 5 | 0.280 8 | 0.367 1 | 0.273 9 | 0.294 7 | 0.301 7 | 0.273 9 | 0.382 3 | 0.176 1 | **** | 0.812 5 | 0.791 7 | 0.734 4 | 0.729 2 | 0.630 2 |
| BZ-13 | 0.367 1 | 0.273 9 | 0.220 5 | 0.337 5 | 0.337 5 | 0.315 9 | 0.240 2 | 0.301 7 | 0.246 9 | 0.445 3 | 0.240 2 | 0.207 6 | **** | 0.875 0 | 0.776 0 | 0.760 4 | 0.651 0 |
| BZ-14 | 0.429 2 | 0.301 7 | 0.273 9 | 0.352 2 | 0.367 1 | 0.359 7 | 0.253 5 | 0.374 7 | 0.315 9 | 0.413 3 | 0.253 5 | 0.233 6 | 0.133 5 | **** | 0.786 5 | 0.750 0 | 0.682 3 |
| BZ-15 | 0.315 9 | 0.240 2 | 0.267 1 | 0.315 9 | 0.287 7 | 0.397 7 | 0.273 9 | 0.323 0 | 0.323 0 | 0.330 2 | 0.315 9 | 0.308 7 | 0.253 5 | 0.240 2 | **** | 0.776 0 | 0.677 1 |
| BZ-16 | 0.367 1 | 0.287 7 | 0.233 6 | 0.294 7 | 0.352 2 | 0.374 7 | 0.308 7 | 0.273 9 | 0.315 9 | 0.413 3 | 0.294 7 | 0.315 9 | 0.273 9 | 0.287 7 | 0.253 5 | **** | 0.682 3 |
| BZ-17 | 0.470 0 | 0.367 1 | 0.413 3 | 0.470 0 | 0.521 3 | 0.530 1 | 0.486 8 | 0.413 3 | 0.548 0 | 0.486 8 | 0.421 2 | 0.461 7 | 0.429 2 | 0.382 3 | 0.390 0 | 0.382 3 | **** |

图4 SCoT标记构建17份白术种质的分子指纹图谱 1-17表示白术种质编号BZ-1-BZ-17;黑色方框代表样品在对应的SCoT引物位点上有扩增条带
Fig. 4 Molecular fingerprint of 17 A. macrocephala germplasms constructed with SCoT markers 1-17 indicate the germplasm numbers BZ-1-BZ-17 of A. macrocephala. The black box indicates the amplification band of the sample at the corresponding SCoT primer site
| [1] | 国家药典委员会. 中华人民共和国药典: 一部[S]. 北京: 中国医药科技出版社, 2020. |
| State Pharmacopoeia Commission Pharmacopoeia of the People's Republic of China: Volume I[S]Beijing: China Pharmaceutical Science and Technology Press, 2020. | |
| [2] |
岳美颖, 潘媛, 敖慧. 白术化学、药理与临床研究进展[J]. 亚太传统医药, 2016, 12(5): 66-68.
doi: 10.11954/ytctyy.201605024 |
| Yue MY, Pan Y, Ao H. Advance on the chemistry, pharmacological and clinical of Atractylodes macrocephala[J]Asia Pac Tradit Med, 2016, 12(5): 66-68. | |
| [3] | 李丽娟. 探讨中药材及中药饮片质量控制对策[J]. 现代诊断与治疗, 2018, 29(17): 2819-2821. |
| Li LJ. Discussion on quality control countermeasures of Chinese herbal medicines and Chinese herbal pieces[J]. Mod Diagn Treat, 2018, 29(17): 2819-2821. | |
| [4] | 刘逸慧, 陈斌龙, 周晓龙, 等. 药用植物白术栽培群体的遗传多样性研究[J]. 中国中药杂志, 2008, 33(23): 2756-2760. |
| Liu YH, Chen BL, Zhou XL, et al. Studies on genetic diversity in cultivated populations of Atractylodes macrocephala[J]. China J Chin Mater Med, 2008, 33(23): 2756-2760. | |
| [5] | 付竹贤, 关仁梅, 王淑敏, 等. 随机扩增多态性DNA分子标记技术在肠球菌分型中的应用[J]. 现代食品科技, 2023, 39(11): 86-93. |
| Fu ZX, Guan RM, Wang SM, et al. Application of random amplified polymorphic DNA molecular marker technique in typing of Enterococcus[J]. Mod Food Sci Technol, 2023, 39(11): 86-93. | |
| [6] | 于彤, 申文辉, 覃日亮, 等. 基于ISSR分子标记技术的土沉香遗传多样性分析[J]. 广西林业科学, 2023, 52(6): 726-732. |
| Yu T, Shen WH, Qin RL, et al. Analysis on genetic diversity of Aquilaria sinensis based on ISSR molecular markers[J]. Guangxi For Sci, 2023, 52(6): 726-732. | |
| [7] | 薛江源, 李艳荣, 孙芮芮, 等. 黄芪的分子标记开发及应用研究进展[J]. 中华中医药杂志, 2023, 38(1): 280-284. |
| Xue JY, Li YR, Sun RR, et al. Research progress on the development and application of molecular markers of Astragali radix[J]. China J Tradit Chin Med Pharm, 2023, 38(1): 280-284. | |
| [8] | 谢薇. 中药材鉴定运用分子标记技术的实践探寻[J]. 光明中医, 2021, 36(19): 3262-3264. |
| Xie W. Exploration on the application of molecular marker technology in the identification of Chinese herbal medicines[J]. Guangming J Chin Med, 2021, 36(19): 3262-3264. | |
| [9] | Collard BCY, MacKill DJ. Start Codon targeted(SCoT)polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants[J]. Plant Mol Biol Report, 2009, 27(1): 86-93. |
| [10] | 周彩霞, 罗培四, 孔方南, 等. 木奶果种质资源的遗传多样性分析及DNA指纹图谱构建[J]. 南方农业学报, 2022, 53(5): 1207-1215. |
| Zhou CX, Luo PS, Kong FN, et al. Genetic diversity analysis and DNA fingerprinting construction with Baccaurea ramiflora Lour. germplasm resources[J]. J South Agric, 2022, 53(5): 1207-1215. | |
| [11] | 王宏利, 赵久成, 赖淼, 等. 基于SCoT分子标记的61份加工型黄瓜种质资源遗传多样性分析及DNA指纹图谱构建[J]. 中国瓜菜, 2024, 37(4): 36-45. |
| Wang HL, Zhao JC, Lai M, et al. Genetic diversity analysis and DNA fingerprinting construction of 61 pro-cessed cucumber germplasm resources based on SCoT markers[J]. China Cucurbits Veg, 2024, 37(4): 36-45. | |
| [12] | Gupta V, Jatav PK, Haq SU, et al. Translation initiation codon(ATG)or SCoT markers-based polymorphism study within and across various Capsicum accessions: insight from their amplification, cross-transferability and genetic diversity[J]. J Genet, 2019, 98(2): 61. |
| [13] | 零唯, 杨丹, 谭勇, 等. 基于SCoT标记的胡椒属药用植物多样性分析[J]. 安徽农业科学, 2023, 51(8): 182-186, 202. |
| Ling W, Yang D, Tan Y, et al. Diversity analysis of Piper medicinal plants based on SCoT markers[J]. J Anhui Agric Sci, 2023, 51(8): 182-186, 202. | |
| [14] | 马玉姣, 何西堂, 吴玉森, 等. 基于SCoT分子标记的37份葡萄种质遗传多样性分析[J]. 中外葡萄与葡萄酒, 2023(4): 18-23. |
| Ma YJ, He XT, Wu YS, et al. Genetic diversity analysis of 37 grape germplasms using SCoT markers[J]. Sino Overseas Grapevine Wine, 2023(4): 18-23. | |
| [15] | 刘新梅, 梁慧珍, 许兰杰, 等. 红花SCoT-PCR反应体系的建立与优化[J]. 分子植物育种, 2022, 20(3): 895-901. |
| Liu XM, Liang HZ, Xu LJ, et al. Establishment and optimization of SCoT-PCR amplification system in saf-flower[J]. Mol Plant Breed, 2022, 20(3): 895-901. | |
| [16] | 邵征绩, 张璐, 郑升, 等. 铁皮石斛SCoT-PCR反应体系的建立及其初步应用[J]. 分子植物育种, 2021, 19(5): 1606-1613. |
| Shao ZJ, Zhang L, Zheng S, et al. Establishment and preliminary application of SCoT-PCR system of dendr-obium officinale kimura et migo[J]. Mol Plant Breed, 2021, 19(5): 1606-1613. | |
| [17] | 尚小红, 严华兵, 曹升, 等. 葛根SCoT-PCR反应体系优化及引物筛选[J]. 南方农业学报, 2018, 49(1): 1-7. |
| Shang XH, Yan HB, Cao S, et al. Optimization of SCoT-PCR reaction system and primer selection for Pueraria DC[J]. J South Agric, 2018, 49(1): 1-7. | |
| [18] | 王瑞娟. 基于ISSR和SCoT分子标记的蒙古黄芪遗传多样性分析[D]. 呼和浩特: 内蒙古大学, 2023. |
| Wang RJ. Genetic diversity analysis of Astragalus mongholicus based on ISSR and SCoT molecular markers[D]. Hohhot: Inner Mongolia University, 2023. | |
| [19] | 瞿印权, 徐雯, 何天友, 等. DNA分子标记技术在竹亚科植物遗传多样性研究中的应用[J]. 安徽农业科学, 2017, 45(34): 141-143. |
| Qu YQ, Xu W, He TY, et al. Application of DNA molecular marker in the study of genetic diversity of bambsoideae[J]. J Anhui Agric Sci, 2017, 45(34): 141-143. | |
| [20] | 张晓峰, 刘福艳. DNA指纹图谱技术在中药研究中的应用进展[J]. 齐鲁药事, 2012, 31(12): 722-725. |
| Zhang XF, Liu FY. Progress in the application of DNA fingerprinting technology in traditional Chinese medicine research[J]. Qilu Pharmaceut Sci, 2012, 31(12): 722-725 | |
| [21] | 李志良, 杜睿琛, 杨杰, 等. 基于ISSR标记的杜鹃花品种DNA指纹图谱的构建[J]. 分子植物育种, 2021, 19(11): 3646-3652. |
| Li ZL, Du RC, Yang J, et al. Construction of DNA fingerprints in different Rhododendron cultivars based on ISSR markers[J]. Mol Plant Breed, 2021, 19(11): 3646-3652. | |
| [22] | 乔菲, 刘杰, 房文亮, 等. 巴戟天饮片及其混伪品的鉴别研究[J]. 药物分析杂志, 2022, 42(4): 668-675. |
| Qiao F, Liu J, Fang WL, et al. Identification study on morindae officinalis Radix decoction pieces and the fakes[J]. Chin J Pharm Anal, 2022, 42(4): 668-675. | |
| [23] | 张安世, 骆扬, 范定臣, 等. 皂荚种质资源SCoT遗传多样性分析及指纹图谱的构建[J]. 广西植物, 2017, 37(11): 1378-1385. |
| Zhang AS, Luo Y, Fan DC, et al. Genetic diversity and fingerprints of Gleditsia sinensis germplasm resource based on SCoT[J]. Guihaia, 2017, 37(11): 1378-1385. | |
| [24] |
潘静, 张俊超, 陈有军, 等. 基于SCoT标记的披碱草属种质遗传多样性分析及指纹图谱构建[J]. 草业学报, 2022, 31(11): 48-60.
doi: 10.11686/cyxb2022137 |
| Pan J, Zhang JC, Chen YJ, et al. Genetic diversity analysis and fingerprint construction of Elymus germplasm based on SCoT markers[J]. Acta Prataculturae Sin, 2022, 31(11): 48-60. | |
| [25] |
Ellegren H, Galtier N. Determinants of genetic diversity[J]. Nat Rev Genet, 2016, 17(7): 422-433.
doi: 10.1038/nrg.2016.58 pmid: 27265362 |
| [26] | 钟明志, 毛莉, 姜媛媛, 等. 基于ISSR和SCoT分子标记的丹参遗传多样性评价及生境因子对丹酚酸和丹参酮的影响[J]. 中草药, 2024, 55(12): 4171-4184. |
| Zhong MZ, Mao L, Jiang YY, et al. Evaluation of genetic diversity of Salvia miltiorrhiza based on ISSR and SCoT molecular markers and the effects of environmental factors on salviolic acid and salvianolic acid[J]. Chinese Materia Medica, 2024, 55(12): 4171-4184. | |
| [27] | 陈虎, 何新华, 罗聪, 等. 龙眼24个品种的SCoT遗传多样性分析[J]. 园艺学报, 2010, 37(10): 1651-1654. |
| Chen H, He XH, Luo C, et al. Analysis on the genetic diversity of 24 longan(Dimocarpus longan)accessions by SCoT markers[J]. Acta Hortic Sin, 2010, 37(10): 1651-1654. | |
| [28] | 高曼熔, 杨天为, 黄诗宇, 等. 广西野生铁皮石斛SCoT体系优化及遗传多样性分析[J/OL]. 分子植物育种, 2024: 1-19. |
| Gao MR, Yang TW, Huang SY, et al. Optimization of SCoT system and genetic diversity analysis of wild Dendrobium officinale in Guangxi[J/OL]. Molecular plant breeding, 2024:1-19. | |
| [29] | 蒋小刚, 王华, 周武先, 等. 基于ISSR分子标记的白术遗传多样性分析[J]. 种子, 2021, 40(4): 6-10. |
| Jiang XG, Wang H, Zhou WX, et al. Genetic diversity analysis of Atractylodes macrocephala based on ISSR markers[J]. Seed, 2021, 40(4): 6-10. | |
| [30] | 蒋小刚, 王华, 张美德. 白术种质资源遗传多样性的SRAP分析[J]. 北方园艺, 2022(1): 109-115. |
| Jiang XG, Wang H, Zhang MD. Genetic diversity of Atractylodes macrocephala by SRAP markers[J]. North Hortic, 2022(1): 109-115. | |
| [31] | 杨珂, 周延清, 段红英, 等. 地黄SCoT分子标记体系的建立和指纹图谱的构建[J]. 广西植物, 2019, 39(5): 608-614. |
| Yang K, Zhou YQ, Duan HY, et al. SCoT molecular marker system and fingerprint in Rehmannia glutinosa[J]. Guihaia, 2019, 39(5): 608-614. | |
| [32] | Xu HY, Li P, Ren GX, et al. Authentication of three source spices of Arnebiae radix using DNA barcoding and HPLC[J]. Front Pharmacol, 2021, 12: 677014. |
| [33] | Zhong YC, Wang HY, Wei QH, et al. Combining DNA barcoding and HPLC fingerprints to trace species of an important traditional Chinese medicine fritillariae Bulbus[J]. Molecules, 2019, 24(18): 3269. |
| [34] |
蒋林峰, 张新全, 黄琳凯, 等. 中国鸭茅主栽品种DNA指纹图谱构建[J]. 植物遗传资源学报, 2014, 15(3): 604-614.
doi: 10.13430/j.cnki.jpgr.2014.03.023 |
| Jiang LF, Zhang XQ, Huang LK, et al. Construction of DNA fingerprinting of dominant orchardgrass(Dactylis glomerata)varieties of China[J]. J Plant Genet Resour, 2014, 15(3): 604-614. | |
| [35] | Verma S, Rana TS. Genetic relationships among wild and cultivated accessions of curry leaf plant(Murraya koenigii(L.) spreng.), as revealed by DNA fingerprinting methods[J]. Mol Biotechnol, 2013, 53(2): 139-149. |
| [36] | Gao P, Ma HY, Luan FS, et al. DNA fingerprinting of Chinese melon provides evidentiary support of seed quality appraisal[J]. PLoS One, 2012, 7(12): e52431. |
| [1] | 谢果珍, 唐圆, 吴仪, 黄莉莉, 谭周进. 七味白术散总苷对菌群失调腹泻小鼠肠道微生物及酶活性的影响[J]. 生物技术通报, 2021, 37(12): 124-131. |
| [2] | 王衍莉, 杨义明, 范书田, 赵滢, 许培磊, 路文鹏, 李昌禹. 基于SSR分子标记的73份山葡萄及杂交后代的遗传多样性分析[J]. 生物技术通报, 2021, 37(1): 189-197. |
| [3] | 李宏博, 庞斌双, 刘丽华, 刘阳娜, 赵昌平, 陈景堂. 河北区试小麦品种(系)DNA指纹图谱构建及遗传差异分析[J]. 生物技术通报, 2015, 31(6): 93-99. |
| [4] | 唐玲;刘平;黄瑛;杨江科;闫云君;. 酵母的分子生物学鉴定[J]. , 2008, 0(05): 84-87. |
| [5] | 张春磊;王爽;王秀利;. 分子生物学技术在水产动物中的研究与应用[J]. , 2006, 0(S1): 209-212. |
| [6] | 秦春圃. 我国藏山羊微卫星DNA复合扩增[J]. , 2003, 0(06): 57-58. |
| [7] | 贾继增. 小麦分子标记研究进展[J]. , 1997, 0(02): 1-5. |
| [8] | 朱遐. 利用DNA指纹图谱检测体克隆变异[J]. , 1995, 0(01): 11-12. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||