








生物技术通报 ›› 2025, Vol. 41 ›› Issue (2): 40-50.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0764
王彤1( ), 肖汉杰1, 谢昊炅1, 张立申2, 姚晓亮3, 严慧1(
), 肖汉杰1, 谢昊炅1, 张立申2, 姚晓亮3, 严慧1( ), 纪守坤1(
), 纪守坤1( )
)
收稿日期:2024-08-08
出版日期:2025-02-26
发布日期:2025-02-28
通讯作者:
严慧,女,副教授,硕士生导师,研究方向 :反刍动物肠道微生态与健康;E-mail: yanhuihui@126.com作者简介:王彤,女,博士研究生,研究方向 :反刍动物营养;E-mail: wang2896784183@163.com
基金资助:
WANG Tong1( ), XIAO Han-jie1, XIE Hao-jiong1, ZHANG Li-shen2, YAO Xiao-liang3, YAN Hui1(
), XIAO Han-jie1, XIE Hao-jiong1, ZHANG Li-shen2, YAO Xiao-liang3, YAN Hui1( ), JI Shou-kun1(
), JI Shou-kun1( )
)
Received:2024-08-08
Published:2025-02-26
Online:2025-02-28
摘要:
蛋白质饲料资源的短缺严重限制着我国畜牧业的持续稳定发展。反刍动物具有将非蛋白氮转化为氨基酸或蛋白质的独特能力,添加非蛋白氮是缓解反刍动物蛋白饲粮短缺的有效途径,但瘤胃氨同化是限制其利用效率的关键步骤。因此,如何提高反刍动物氮同化效率是缓解蛋白饲粮短缺的重要手段。氨同化指游离氨并入碳骨架形成含氮有机物的过程。瘤胃微生物可以将游离的氨同化为微生物蛋白质(microbial protein, MCP),为反刍动物提供49%-98%的可代谢蛋白质。瘤胃氨同化作用可通过4条通路进行:①谷氨酸脱氢酶(glutamate dehydrogenase, GDH):氨与α-酮戊二酸在GDH的作用下发生胺化生成谷氨酸;②谷氨酰胺合成酶-谷氨酸合成酶(glutamine synthetase-glutamate synthase, GS-GOGAT):氨和谷氨酸经GS催化后形成谷氨酰胺;③丙氨酸脱氢酶(alanine dehydrogenase, ADH):氨和丙酮酸可经过ADH通路生成丙氨酸;④天冬酰胺合成酶(asparagine synthetase, AS):天冬氨酸和氨可通过AS通路发生酰胺化生成天冬酰胺。本文深入探究了瘤胃微生物氨同化通路反应过程,综述了瘤胃4条氨同化通路过程及其关键微生物、酶和影响因素,并通过构建系统发育树解析了瘤胃细菌不同氨同化基因的进化关系,为反刍动物氨氮高效利用提供理论支持。
王彤, 肖汉杰, 谢昊炅, 张立申, 姚晓亮, 严慧, 纪守坤. 瘤胃氨同化通路及基因进化研究进展[J]. 生物技术通报, 2025, 41(2): 40-50.
WANG Tong, XIAO Han-jie, XIE Hao-jiong, ZHANG Li-shen, YAO Xiao-liang, YAN Hui, JI Shou-kun. Research Progress in Ammonia Assimilation and Genetic Evolution in Rumen[J]. Biotechnology Bulletin, 2025, 41(2): 40-50.
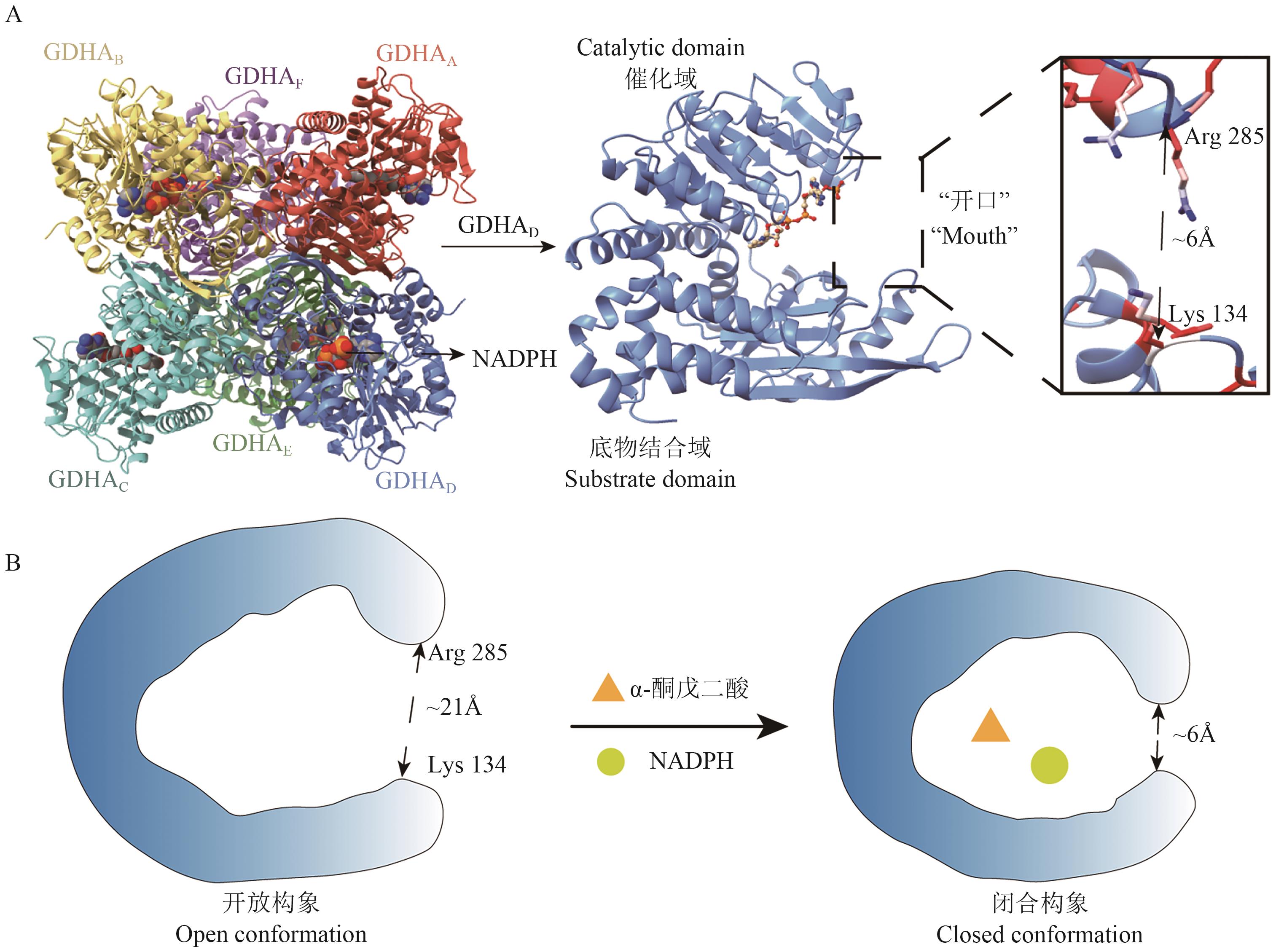
图2 GDH结构图A:GDH六聚体和亚基结构示意图,基于Butyrivibrio fibrisolvens全基因组数据(NCBI序列号:GCA_037113525.1),利用AlphaFold3平台构建;B:底物与辅酶结合后GDH构象变化示意图
Fig. 2 Structure of GDHA: Cartoon representation of hexameric and subunit GDH structure. The structure is built based on the whole genome of B. fibrisolvens using the AlphaFold3 platform (NCBI GenBank: GCA_037113525.1). B: Schematic diagram depicting conformational change in GDH structure upon substrate and coenzyme binding
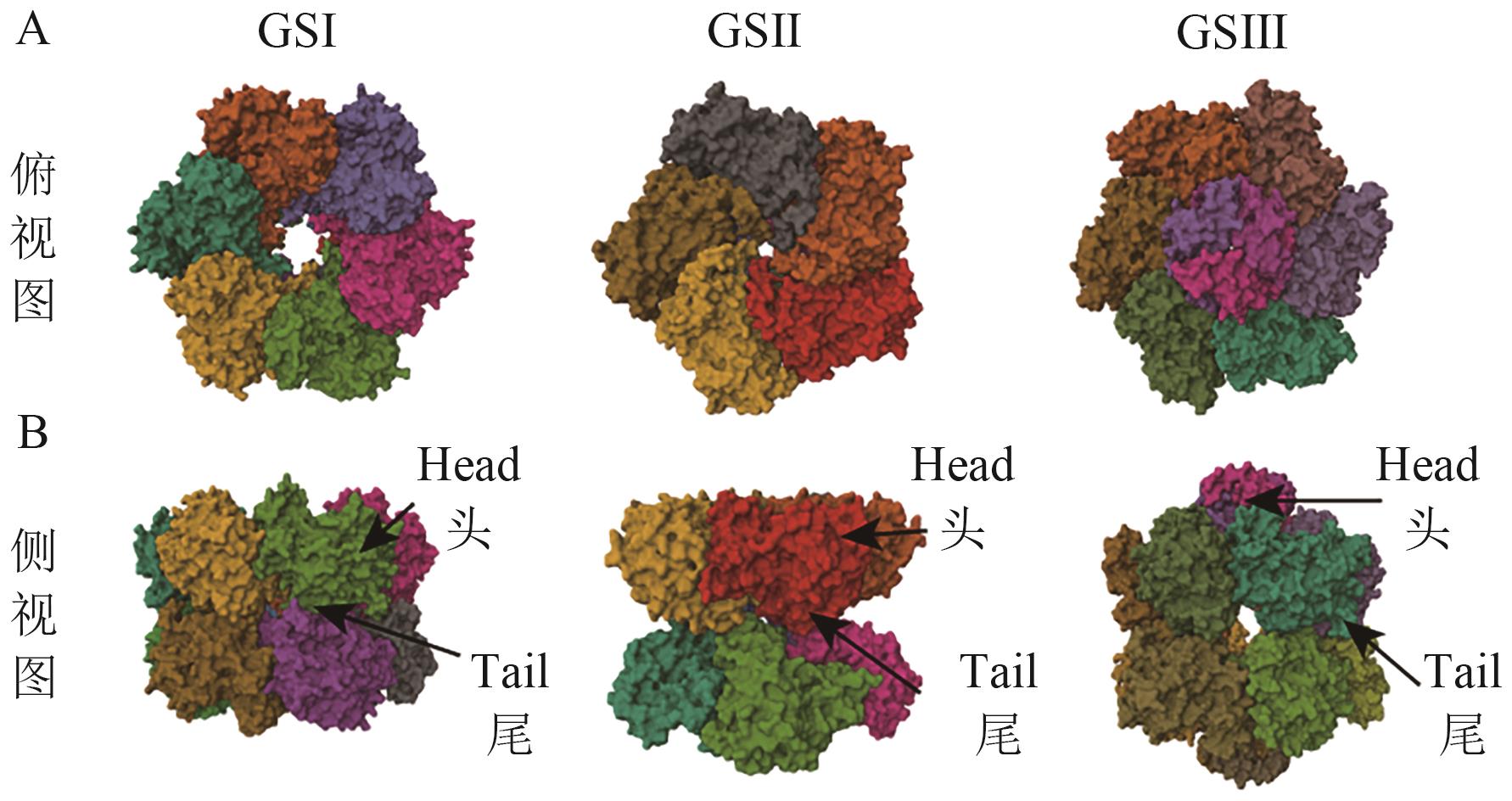
图4 三类GS结构图来自PDB数据库,序列号分别为5ZLP、7v4i、8TGE
Fig. 4 Multimeric structures of the three types of GSThe structures are from the PDB database, with serial number 5ZLP, 7v4i, and 8TGE, respectively
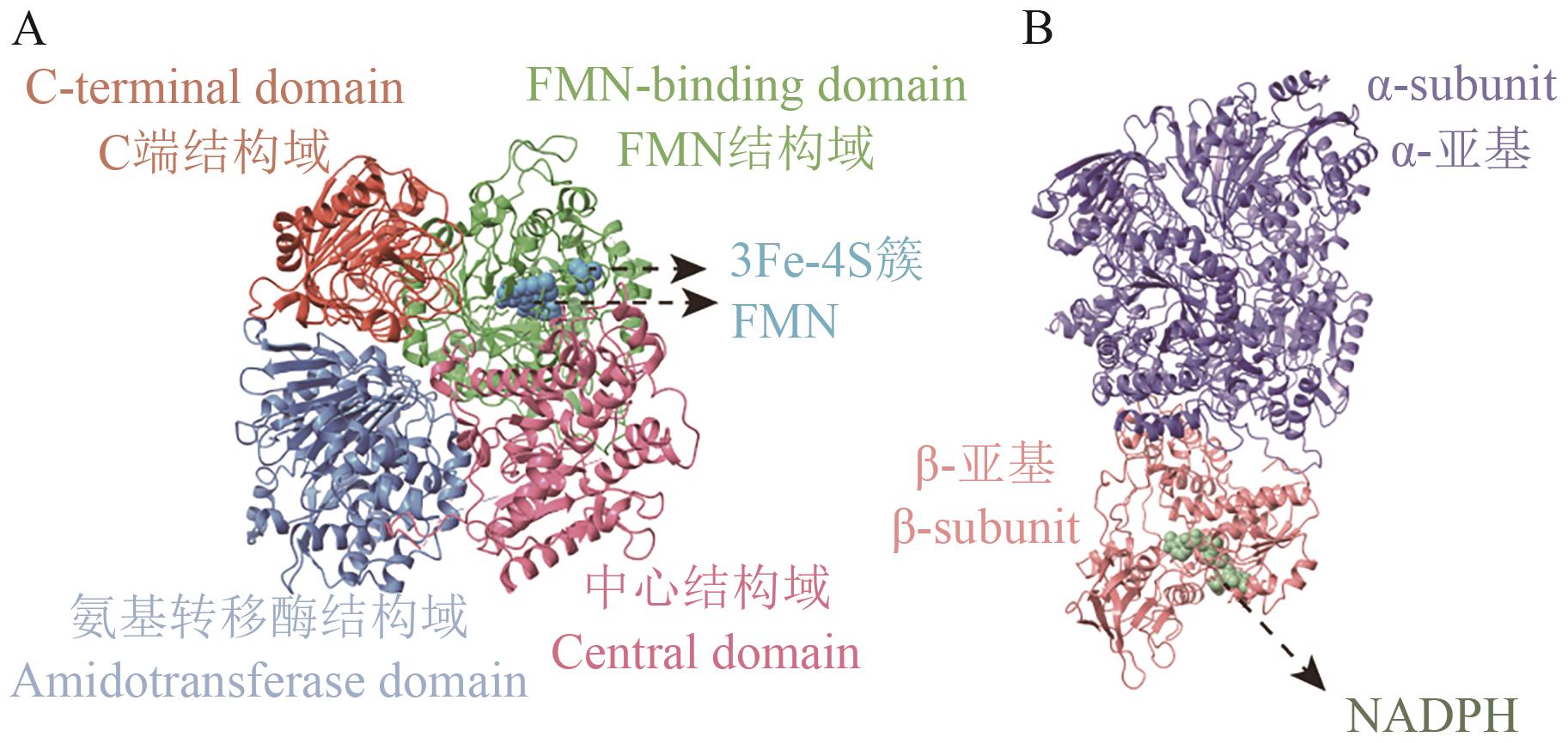
图5 GOGAT结构图A:Fd-GOGAT结构示意图,结构源于PDB(1LLZ);B:NADPH-GOGAT结构示意图,基于B. fibrisolvens(NCBI GenBank: GCA_037113525.1)全基因组数据,利用AlphaFold3平台预测
Fig. 5 Structure of GOGATA: Schematic diagram of Fd-GOGAT structure, from PDB (1 LLZ). B: Schematic diagram of NADPH-GOGAT structure. Structure based on B. fibrisolvens genome data (NCBI GenBank: GCA_037113525.1), predicted by AlphaFold3
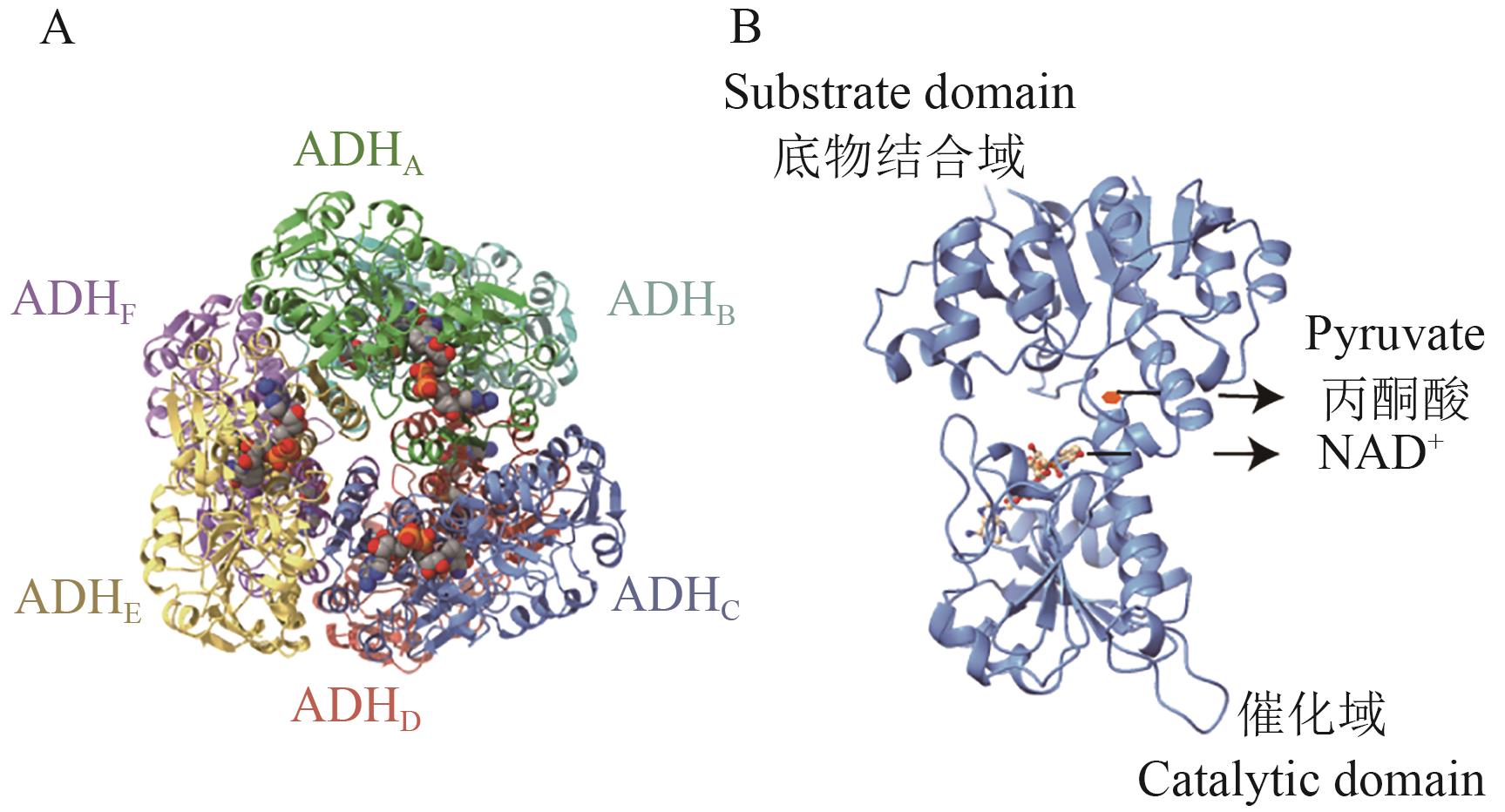
图7 ADH结构图A:ADH六聚体结构图,不同颜色代表不同亚基;B:单个亚基结构示意图,其中球棒状模型代表NAD+,红色六边形代表丙酮酸。图中所有结构均以B. fragilis全基因组数据为基础,利用AlphaFold3平台预测蛋白结构(NCBI序列号:GCA_016889925.1)
Fig. 7 Structure of ADHA: The structural diagram of ADH hexamer, with different colors representing different subunits. B: The structural diagram of ADH monomer, where the ball-and-stick model indicates NAD+ and the red hexagon indicates pyruvate. The structure in the figure is based on the whole genome data of B. fragilis, and protein structure prediction was conducted using the AlphaFold3 platform (NCBI GenBank: GCA_016889925.1)
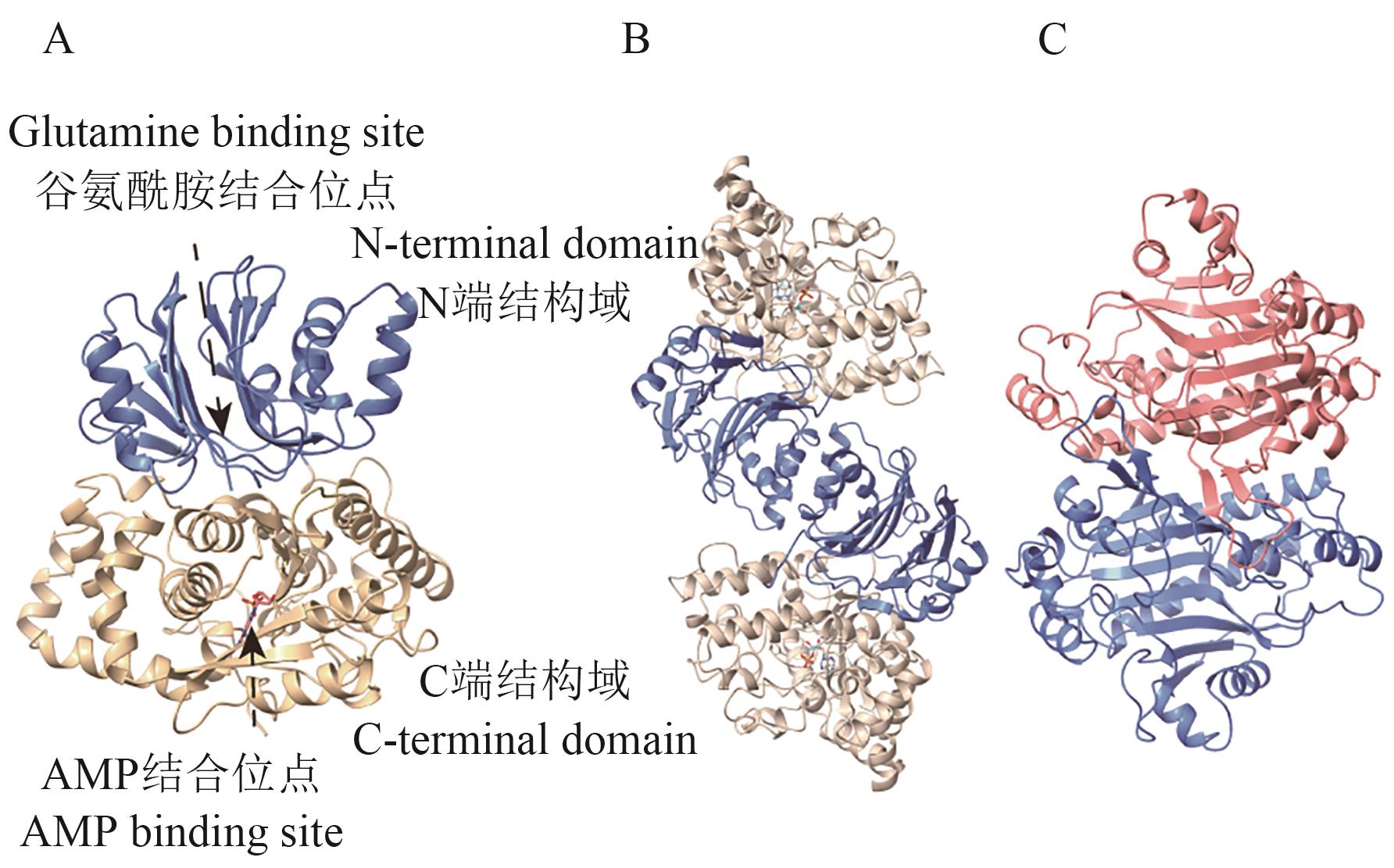
图9 AS结构图A:AS-B单个亚基结构图;B:AS-B二聚体形式结构图;C:AS-A二聚体形式结构图。图中结构均基于B. fibrisolvens全基因组数据(NCBI GenBank:GCA_037113525.1),利用AlphaFold3平台构建
Fig. 9 Structure of ASA: The structural diagram of AS-B monomer. B: Structure diagram of AS-B dimer. C: Structure diagram of AS-A dimer. The structures are all built based on the whole genome data of B. fibrisolvens (NCBI GenBank: GCA_037113525.1) using the AlphaFold3 platform
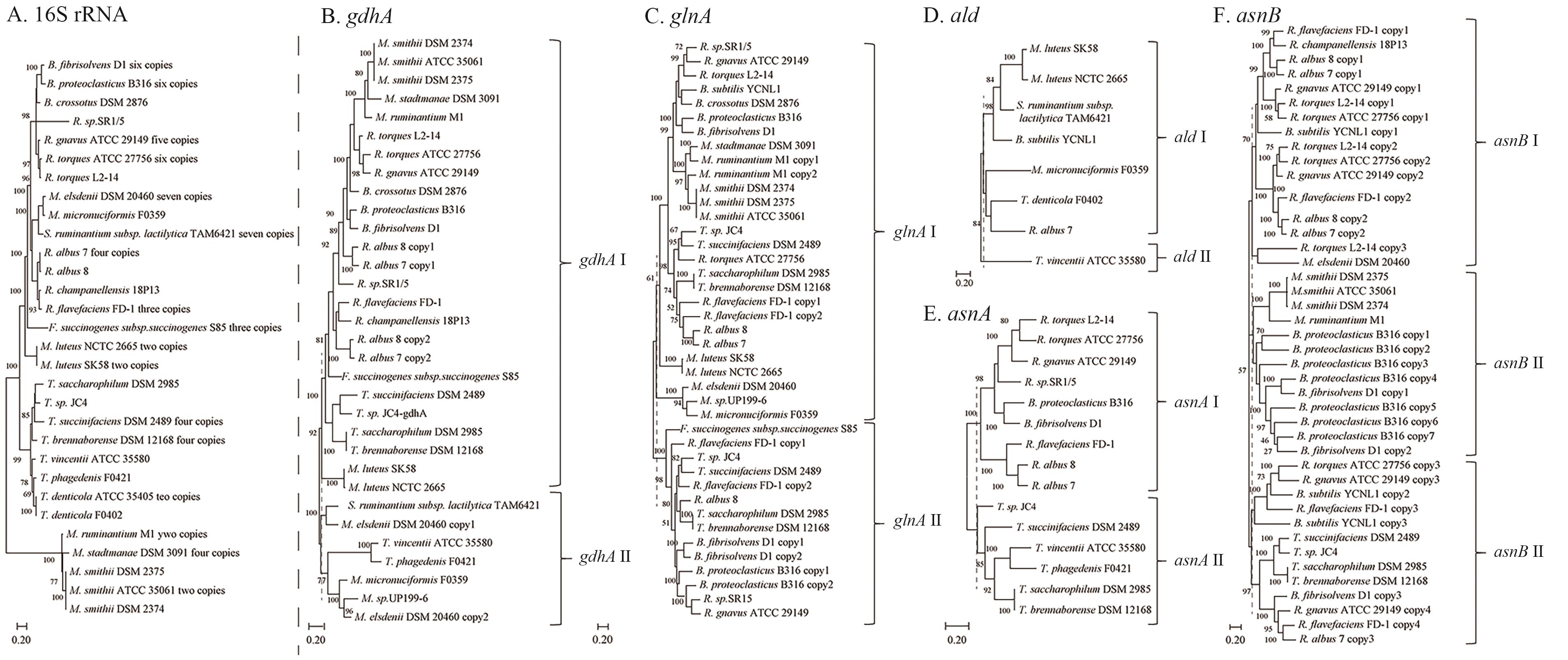
图11 16SrRNA和氨同化通路关键基因系统发育树A:31株瘤胃细菌的16S rRNA系统发育树;B:gdhA基因系统发育树(GDH通路);C:glnA基因系统发育树(GS-GOGAT通路);D:ald基因系统发育树(ADH通路);E和F分别为asnA和asnB基因系统发育树(AS通路)。B-F中的虚线为进化树分支的基因分型位置。基于HMMER检索同源基因,使用MEGA 11 最大似然法构建进化树,bootstrap值设为100
Fig. 11 Phylogenetic trees of 16S rRNA and key genes in the ammonia assimilation pathwayA: 16S rRNA phylogenetic tree of 31 rumen bacterial strains. B: Phylogenetic tree of gdhA gene (GDH pathway). C: Phylogenetic trees of glnA gene (GS-GOGAT pathway). D: Phylogenetic tree of ald gene (ADH pathway); E and F is phylogenetic trees of asnA and asnB genes, respectively (AS pathway). The dashed lines in Fig. B-F indicate the location of genotyping. Homologous genes were retrieved using HMMER. Trees are constructed using MEGA 11 with maximum likelihood method. Bootstrap values are 100
| 1 | 刁其玉, 张春桃. 反刍动物营养与日粮中的蛋白替代技术策略 [J]. 饲料工业, 2024, 45(2): 1-7. |
| Diao QY, Zhang CT. Nutrition of ruminants and protein substitution strategies in diets [J]. Feed Ind, 2024, 45(2): 1-7. | |
| 2 | 王芃芃, 谭支良. 反刍动物瘤胃微生物氨同化作用研究进展 [J]. 动物营养学报, 2010, 22(5): 1171-1176. |
| Wang PP, Tan ZL. Research advances in characterisation of ammonia-assimilation of microorganisms in the rumen [J]. Chin J Anim Nutr, 2010, 22(5): 1171-1176. | |
| 3 | Alemneh T. Urea metabolism and recycling in ruminants [J]. Biomed J Sci Tech Res, 2019, 20(1): 14790-14796. |
| 4 | Patra AK, Aschenbach JR. Ureases in the gastrointestinal tracts of ruminant and monogastric animals and their implication in urea-N/ammonia metabolism: a review [J]. J Adv Res, 2018, 13: 39-50. |
| 5 | Pengpeng W, Tan ZL. Ammonia assimilation in rumen bacteria: a review [J]. Anim Biotechnol, 2013, 24(2): 107-128. |
| 6 | Xiao SS, Zhang HD, Zhu RK, et al. Ammonia reduction by the gdhA and glnA genes from bacteria in laying hens [J]. Ecotoxicol Environ Saf, 2021, 222: 112486. |
| 7 | Zurak D, Kljak K, Aladrović J. Metabolism and utilisation of non-protein nitrogen compounds in ruminants: a review [J]. J Cent Eur Agric, 2023, 24(1): 1-14. |
| 8 | Geron LJV, Garcia J, de Aguiar SC, et al. Effect of slow release urea in sheep feed on nitrogen balance [J]. Semina Ciências Agrárias, 2018, 39(2): 683. |
| 9 | Cardoso GS, Pereira LB, Martini AP, et al. Non-carcass components of finished feedlot steers fed with slow-release or agricultural urea in substitution of soybean meal [J]. Semina Cienc Agrar, 2018, 39: 2761-2770. |
| 10 | Zhong CL, Long RJ, Stewart GS. The role of rumen epithelial urea transport proteins in urea nitrogen salvage: a review [J]. Anim Nutr, 2022, 9: 304-313. |
| 11 | 德吉, 郑琳, 吴玉江, 等. 缓释尿素对藏西北绒山羊生长性能、氮代谢、血浆指标和瘤胃发酵的影响[J]. 动物营养学报, 2023, 35(10): 6526-6537. |
| De J, Zheng L, Wu YJ, et al. Effects of slow-release urea on growth performance, nitrogen metabolism, blood indices and rumen fermentation of northwestern Tibetan Cashmere Goats[J]. Chin J Anim Nutr, 2023, 35(10): 6526-6537. | |
| 12 | Zhang ZB, Sun YQ, Zhong XH, et al. Dietary crude protein and protein solubility manipulation enhances intestinal nitrogen absorption and mitigates reactive nitrogen emissions through gut microbiota and metabolome reprogramming in sheep [J]. Anim Nutr, 2024, 18: 57-71. |
| 13 | Zhang J, Zheng N, Shen WJ, et al. Synchrony degree of dietary energy and nitrogen release influences microbial community, fermentation, and protein synthesis in a rumen simulation system [J]. Microorganisms, 2020, 8(2): 231. |
| 14 | Xue FG, Nan XM, Li YL, et al. Metagenomic insights into effects of thiamine supplementation on ruminal non-methanogen Archaea in high-concentrate diets feeding dairy cows [J]. BMC Vet Res, 2019, 15(1): 7. |
| 15 | Wang PP, Tan ZL, Guan LL, et al. Ammonia and amino acids modulates enzymes associated with ammonia assimilation pathway by ruminal microbiota in vitro [J]. Livest Sci, 2015, 178: 130-139. |
| 16 | Singh S, Koli P, Bhadoria BK, et al. Proanthocyanidins modulate rumen enzyme activities and protein utilization in vitro [J]. Molecules, 2022, 27(18): 5870. |
| 17 | Koli P, Singh S, Bhadoria BK, et al. Sequential extraction of proanthocyanidin fractions from Ficus species and their effects on rumen enzyme activities in vitro [J]. Molecules, 2022, 27(16): 5153. |
| 18 | 王天武, 王腾达, 张玉晶, 等. 非蛋白氮在反刍动物生产中的研究进展 [J]. 饲料研究, 2023, 46(7): 138-141. |
| Wang TW, Wang TD, Zhang YJ, et al. Progress of research on non-protein nitrogen in ruminant production [J]. Feed Res, 2023, 46(7): 138-141. | |
| 19 | Flores PAM, Correa-Llantén DN, Blamey JM. A thermophilic microorganism from Deception Island, Antarctica with a thermostable glutamate dehydrogenase activity [J]. Biol Res, 2018, 51(1): 55. |
| 20 | 武文慧. 枯草芽孢杆菌来源谷氨酸脱氢酶的分子改造及其应用 [D]. 无锡: 江南大学, 2022. |
| Wu WH. Molecular modification of glutamate dehydrogenase from Bacillus subtilis and its application [D]. Wuxi: Jiangnan University, 2022. | |
| 21 | 陆利兵. 谷氨酸脱氢酶的筛选、表达及改造 [D]. 杭州: 浙江大学, 2020. |
| Lu LB. Screening, expression and modification of glutamate dehydrogenases [D]. Hangzhou: Zhejiang University, 2020. | |
| 22 | 田光明. 地衣芽胞杆菌谷氨酸脱氢酶和聚γ-谷氨酸降解酶的研究 [D]. 武汉: 华中农业大学, 2014. |
| Tian GM. Study of glutamate dehydrogenase and poly-γ-glutamic acid hydrolase in Bacillus licheniformis [D]. Wuhan: Huazhong Agricultural University, 2014. | |
| 23 | 尹新坚. 谷氨酸脱氢酶的挖掘、改造及其在L-草铵膦制备中的应用 [D]. 杭州: 浙江大学, 2019. |
| Yin XJ. Mining, engineering and application of glutamate dehydrogenases for enzymatic preparation of L-phosphinothricin [D]. Hangzhou: Zhejiang University, 2019. | |
| 24 | Prakash P, Punekar NS, Bhaumik P. Structural basis for the catalytic mechanism and α-ketoglutarate cooperativity of glutamate dehydrogenase [J]. J Biol Chem, 2018, 293(17): 6241-6258. |
| 25 | Godsora BKJ, Prakash P, Punekar NS, et al. Molecular insights into the inhibition of glutamate dehydrogenase by the dicarboxylic acid metabolites [J]. Proteins, 2022, 90(3): 810-823. |
| 26 | 陈子晗. 生物催化α-酮戊二酸为谷氨酸 [D]. 杭州: 浙江大学, 2019. |
| Chen ZH. Biocatalysis of α-ketoglutaric acid to glutamic acid [D]. Hangzhou: Zhejiang University, 2019. | |
| 27 | Li CD, Zhao TJ, Ren LY, et al. Characterization of a novel glutamate dehydrogenase gene and its response to heat stress in the sea urchin Strongylocentrotus intermedius [J]. Aquac Rep, 2023, 28: 101446. |
| 28 | Ando T, Sugawara Y, Nishio R, et al. Cloning and characterization of the glutamate dehydrogenase gene in Streptococcus bovis [J]. Anim Sci J, 2017, 88(7): 1027-1033. |
| 29 | Mara P, Fragiadakis GS, Gkountromichos F, et al. The pleiotropic effects of the glutamate dehydrogenase (GDH) pathway in Saccharomyces cerevisiae [J]. Microb Cell Fact, 2018, 17(1): 170. |
| 30 | Kim JN, Henriksen ED, Cann IKO, et al. Nitrogen utilization and metabolism in Ruminococcus albus 8 [J]. Appl Environ Microbiol, 2014, 80(10): 3095-3102. |
| 31 | Antonopoulos DA, Aminov RI, Duncan PA, et al. Characterization of the gene encoding glutamate dehydrogenase (gdhA) from the ruminal bacterium Ruminococcus flavefaciens FD-1 [J]. Arch Microbiol, 2003, 179(3): 184-190. |
| 32 | Janssen DB, op den Camp HJ, Leenen PJ, et al. The enzymes of the ammonia assimilation in Pseudomonas aeruginosa [J]. Arch Microbiol, 1980, 124(2/3): 197-203. |
| 33 | Kim JN, Méndez-García C, Geier RR, et al. Metabolic networks for nitrogen utilization in Prevotella ruminicola 23 [J]. Sci Rep, 2017, 7(1): 7851. |
| 34 | Kimura T, Kobayashi K. Role of glutamate synthase in biofilm formation by Bacillus subtilis [J]. J Bacteriol, 2020, 202(14): e00120-20. |
| 35 | Guo J, Man ZW, Rao ZM, et al. Improvement of the ammonia assimilation for enhancing L-arginine production of Corynebacterium crenatum [J]. J Ind Microbiol Biotechnol, 2017, 44(3): 443-451. |
| 36 | Zheng Y, Cabassa-Hourton C, Planchais S, et al. The proline cycle as an eukaryotic redox valve [J]. J Exp Bot, 2021, 72(20): 6856-6866. |
| 37 | de Carvalho Fernandes G, Turchetto-Zolet AC, Pereira Passaglia LM. Glutamine synthetase evolutionary history revisited: tracing back beyond the last universal common ancestor [J]. Evolution, 2022, 76(3): 605-622. |
| 38 | 韩英坤. 巨大芽孢杆菌N6谷氨酰胺合成酶酶学性质的研究和改良 [D]. 武汉: 华中农业大学, 2017. |
| Han YK. Study and improvement on enzymatic properties of glutamine synthetase from Bacillus megaterium N6 [D]. Wuhan: Huazhong Agricultural University, 2017. | |
| 39 | Travis BA, Peck JV, Salinas R, et al. Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria [J]. Nat Commun, 2022, 13(1): 3793. |
| 40 | 刘川顺. 谷氨酰胺合成酶产酶菌株的筛选 [D]. 合肥: 安徽农业大学, 2010. |
| Liu CS. Screening of glutamine synthase producing strain [D]. Hefei: Anhui Agricultural University, 2010. | |
| 41 | dos Santos Moreira CD, Ramos MJRN, Fernandes PMAA. Glutamine synthetase structure-catalysis relationship—recent advances and applications [J]. WIREs Comput Mol Sci, 2019, 9(4): e1399. |
| 42 | Joo HK, Park YW, Jang YY, et al. Structural analysis of glutamine synthetase from Helicobacter pylori [J]. Sci Rep, 2018, 8(1): 11657. |
| 43 | Matheron C, Delort AM, Gaudet G, et al. Interactions between carbon and nitrogen metabolism in Fibrobacter succinogenes S85: a 1H and 13C nuclear magnetic resonance and enzymatic study [J]. Appl Environ Microbiol, 1999, 65(5): 1941-1948. |
| 44 | Fortunato S, Nigro D, Lasorella C, et al. The role of glutamine synthetase (GS) and glutamate synthase (GOGAT) in the improvement of nitrogen use efficiency in cereals [J]. Biomolecules, 2023, 13(12): 1771. |
| 45 | van den Heuvel RHH, Svergun DI, Petoukhov MV, et al. The active conformation of glutamate synthase and its binding to ferredoxin [J]. J Mol Biol, 2003, 330(1): 113-128. |
| 46 | Suzuki A. Glutamate synthase and amino acid synthesis in higher plants [M]//Advances in Botanical Research. Amsterdam: Elsevier, 2021: 129-144. |
| 47 | Gu PF, Ma QQ, Zhao S, et al. Alanine dehydrogenases from four different microorganisms: characterization and their application in L-alanine production [J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 123. |
| 48 | 李嫣妍. 枯草芽孢杆菌丙氨酸脱氢酶的重组表达及理性设计 [D]. 大连: 大连理工大学, 2018. |
| Li YY. Recombinant expression and rational design of alanine dehydrogenase from Bacillus subtilis [D]. Dalian: Dalian University of Technology, 2018. | |
| 49 | 胡小祥. 极光螺杆菌丙氨酸脱氢酶的酶学性质研究、改造及应用[D]. 江南:江南大学,2021. |
| Hu XX. Enzymatic properties,direction evolution and application of alanine dehydrogenase from Helicobacter aurati[D]. Jiangnan: Jiangnan University, 2021. | |
| 50 | Erfle JD, Sauer FS, Mahadevan S. Effect of ammonia concentration on activity of enzymes of ammonia assimilation and on synthesis of amino acids by mixed rumen bacteria in continuous culture [J]. J Dairy Sci, 1977, 60(7): 1064-1072. |
| 51 | Dave UC, Kadeppagari RK. Alanine dehydrogenase and its applications-A review [J]. Crit Rev Biotechnol, 2019, 39(5): 648-664. |
| 52 | Dedeakayoğulları H, Valjakka J, Turunen O, et al. Application of reductive amination by heterologously expressed Thermomicrobium roseum L-alanine dehydrogenase to synthesize L-alanine derivatives [J]. Enzyme Microb Technol, 2023, 169: 110265. |
| 53 | Jeong JA, Oh JI. Alanine dehydrogenases in mycobacteria [J]. J Microbiol, 2019, 57(2): 81-92. |
| 54 | 程维舜, 任俭, 施先锋, 等. 天冬酰胺合成酶的特性及其功能研究进展 [J]. 安徽农业科学, 2013, 41(3): 948-950, 956. |
| Cheng WS, Ren J, Shi XF, et al. Research progress on asparagine synthetase characteristics and function [J]. J Anhui Agric Sci, 2013, 41(3): 948-950, 956. | |
| 55 | Lomelino CL, Andring JT, McKenna R, et al. Asparagine synthetase: Function, structure, and role in disease [J]. J Biol Chem, 2017, 292(49): 19952-19958. |
| 56 | Li Q, Zhang HW, Song Y, et al. Alanine synthesized by alanine dehydrogenase enables ammonium-tolerant nitrogen fixation in Paenibacillus sabinae T27 [J]. Proc Natl Acad Sci USA, 2022, 119(49): e2215855119. |
| 57 | Liu YP, Li LX, Wang JD, et al. Enhancing glutamine production by optimising the GS-GOGAT pathway in Corynebacterium glutamicum and NH4 +-limited fermentation [J]. Auth Pre, 2021 |
| 58 | Kim SH, Kim BG. NAD(+)-specific glutamate dehydrogenase (EC.1.4.1.2) in Streptomyces coelicolor; in vivo characterization and the implication for nutrient-dependent secondary metabolism [J]. Appl Microbiol Biotechnol, 2016, 100(12): 5527-5536. |
| 59 | Feng KX, Wang W, Rong JS, et al. Construction of recombinant Pichia pastoris strains for ammonia reduction by the gdhA and glnA regulatory genes in laying hens [J]. Ecotoxicol Environ Saf, 2022, 234: 113376. |
| 60 | Fu WL, Duan PF, Wang Q, et al. Transcriptomics reveals the effect of ammonia nitrogen concentration on Pseudomonas stutzeri F2 assimilation and the analysis of amtB function [J]. Synth Syst Biotechnol, 2023, 8(2): 262-272. |
| 61 | Ertan H. The effect of various culture conditions on the levels of ammonia assimilatory enzymes of Corynebacterium callunae [J]. Arch Microbiol, 1992, 158(1): 42-47. |
| 62 | Yamamoto I, Abe A, Saito H, et al. The pathway of ammonia assimilation in Bacteroides fragilis [J]. J Gen Appl Microbiol, 1984, 30(6): 499-508. |
| 63 | Wen ZT, Peng LS, Morrison M. The glutamine synthetase of Prevotella bryantii B14 is a family III enzyme (GlnN) and glutamine supports growth of mutants lacking glutamate dehydrogenase activity [J]. FEMS Microbiol Lett, 2003, 229(1): 15-21. |
| 64 | Tesch M, Aa DG, Sahm H. In vivo fluxes in the ammonium-assimilatory pathways in Corynebacterium glutamicum studied by 15N nuclear magnetic resonance [J]. Appl Environ Microbiol, 1999, 65(3): 1099-1109. |
| 65 | 吴优. Arthrobacter ureafaciens CZ31氨氮降解特性及氨同化关键酶的研究 [D]. 武汉: 武汉科技大学, 2018. |
| Wu Y. Study on degradation characteristics of ammonia nitrogen and key enzymes of ammonia assimilation in Arthur Ureafaciens C31 [D]. Wuhan: Wuhan University of Science and Technology, 2018. | |
| 66 | Sun YW, Zhang YB, Zhao TT, et al. Acetylation coordinates the crosstalk between carbon metabolism and ammonium assimilation in Salmonella enterica [J]. EMBO J, 2023, 42(13): e112333. |
| 67 | Wang W, Lu WY, Papadakis VG, et al. Microbial protein production under mixotrophic mode: ammonium assimilation pathway and C/N ratio optimization [J]. J Environ Chem Eng, 2024, 12(1): 111727. |
| 68 | Zhong ZK, Wang C, Zhang HD, et al. Sodium butyrate reduces ammonia emissions through glutamate metabolic pathways in cecal microorganisms of laying hens [J]. Ecotoxicol Environ Saf, 2022, 233: 113299. |
| 69 | Hailemariam S, Zhao SG, Wang JQ. Complete genome sequencing and transcriptome analysis of nitrogen metabolism of Succinivibrio dextrinosolvens strain Z6 isolated from dairy cow rumen [J]. Front Microbiol, 2020, 11: 1826. |
| [1] | 王璐, 刘理想, 孙大明, 刘军花. 丁酸调控幼龄反刍动物瘤胃上皮发育研究进展[J]. 生物技术通报, 2020, 36(2): 49-57. |
| [2] | 朱遐. FAO检测山羊的病毒抗性[J]. , 1996, 0(05): 23-24. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||