








生物技术通报 ›› 2022, Vol. 38 ›› Issue (7): 186-193.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1072
于秋琳1( ), 马婧怡1, 赵盼1, 孙鹏芳1, 何玉美1, 刘世彪2, 郭惠红1(
), 马婧怡1, 赵盼1, 孙鹏芳1, 何玉美1, 刘世彪2, 郭惠红1( )
)
收稿日期:2021-08-20
出版日期:2022-07-26
发布日期:2022-08-09
作者简介:于秋琳,女,硕士研究生,研究方向:植物发育生物学;E-mail: 基金资助:
YU Qiu-lin1( ), MA Jing-yi1, ZHAO Pan1, SUN Peng-fang1, HE Yu-mei1, LIU Shi-biao2, GUO Hui-hong1(
), MA Jing-yi1, ZHAO Pan1, SUN Peng-fang1, HE Yu-mei1, LIU Shi-biao2, GUO Hui-hong1( )
)
Received:2021-08-20
Published:2022-07-26
Online:2022-08-09
摘要:
本研究旨在深入了解药用植物绞股蓝地上茎向根状茎转变过程中两个重要的miRNAs(GpmiR156a和GpmiR166b)在调控植物生长发育方面的作用。采用同源克隆的方法获得GpMIR156a和GpMIR166b基因,进行了生物信息学分析。通过构建植物过表达载体并成功转化拟南芥,研究GpMIR156a和GpMIR166b的过表达对植株表型的影响。GpMIR156a过表达导致拟南芥幼苗叶片数量和分枝增多,并加速了种子萌发、促进根的伸长,表明GpmiR156a能够促进营养器官发育和种子萌发,但抑制童期向成熟期发育的转变。然而,GpMIR166b过表达导致拟南芥植株矮小及早熟性状即花序茎上毛状体数量增多,并延缓种子的萌发,但促进根伸长,表明GpmiR166b在一定程度上影响营养器官的发育和延缓种子萌发,但能促进童期向成熟期发育的转变。研究结果拓宽了miR156a和miR166b在调控植物生长发育方面的认识。
于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193.
YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b[J]. Biotechnology Bulletin, 2022, 38(7): 186-193.
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| GpmiR156a-F | TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-R | GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-F | CAGATACATGTACGAGGWGTTCA |
| GpmiR166b-R | GCTTCATCATYAYACCAATCTGC |
| GpmiR156a-Ft | TGGAGAGAACACGGGGGACTCTAGA TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-Rt | TAACATAAGGGACTGACCACCCGGG GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-Ft | TGGAGAGAACACGGGGGACTCTAGA CAGATA- CATGTACGAGGWGTTCA |
| GpmiR166b-Rt | TAACATAAGGGACTGACCACCCGGG GCTTCA- TCATYAYACCAATCTGC |
| p121-F | CGTCTTCAAAGCAAGTGGATT |
| p121-R | CCAACGCTGATCAATTCCAC |
表1 绞股蓝GpmiR156a和GpmiR166b的引物信息
Table 1 Primer information of G. pentaphyllu GpmiR156a and GpmiR166b
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| GpmiR156a-F | TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-R | GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-F | CAGATACATGTACGAGGWGTTCA |
| GpmiR166b-R | GCTTCATCATYAYACCAATCTGC |
| GpmiR156a-Ft | TGGAGAGAACACGGGGGACTCTAGA TCTTYACWCAACWTCTCACCCA |
| GpmiR156a-Rt | TAACATAAGGGACTGACCACCCGGG GAGARAYCWGCATAWCTCAATMACC |
| GpmiR166b-Ft | TGGAGAGAACACGGGGGACTCTAGA CAGATA- CATGTACGAGGWGTTCA |
| GpmiR166b-Rt | TAACATAAGGGACTGACCACCCGGG GCTTCA- TCATYAYACCAATCTGC |
| p121-F | CGTCTTCAAAGCAAGTGGATT |
| p121-R | CCAACGCTGATCAATTCCAC |

图2 绞股蓝GpmiR156a和GpmiR166b的前体序列 A:GpmiR156a前体序列;B:GpmiR166b前体序列;下划线指示GpmiR156a和GpmiR166b的成熟体
Fig. 2 Precursor sequences of G. pentaphyllu GpmiR156a and GpmiR166b A:GpmiR156a precursor sequence. B:GpmiR166b precursor sequence. Underlines indicate mature sequences of GpmiR156a and GpmiR166b,respectively

图3 绞股蓝GpmiR156a和GpmiR166b的二级结构 A:GpmiR156a的二级结构;B:GpmiR166b的二级结构;图中颜色的变化代表碱基配对概率,红色代表概率最大
Fig. 3 Secondary structures of G. pentaphyllu GpmiR156a and GpmiR166b A:Secondary structure of GpmiR156a. B:Secondary structure of GpmiR166b. The color change in the figure refers to the base pairing probability and the red ones refer to the highest probability
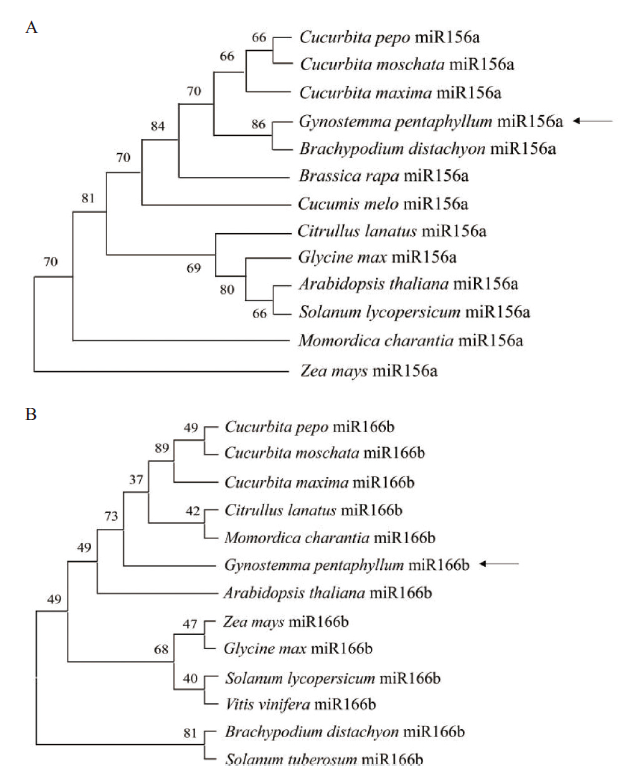
图4 绞股蓝GpmiR156a和GpmiR166b的进化分析 A:GpmiR156a的进化分析;B:GpmiR166b的进化分析;Gynostemma pentaphyllum:绞股蓝;Arabidopsis thaliana:拟南芥;Brassica rapa:芜菁;Zea mays:玉米;Glycine max:大豆;Solanum lycopersicum:番茄;Brachypodium distachyon:二穗短柄草;Cucumis melo:甜瓜;Citrullus lanatus:西瓜;Momordica charantia:苦瓜;Cucurbita pepo:西葫芦;Cucurbita maxima:笋瓜;Cucurbita moschata:南瓜;Vitis vinifera:葡萄;Solanum tuberosum:马铃薯;箭头指示GpmiR156a和GpmiR166b的位置
Fig. 4 Phylogenetic analysis of G. pentaphyllu GpmiR156a and GpmiR166b A:Phylogenetic analysis of GpmiR156a. B:Phylogenetic analysis of GpmiR166b. Arrows indicate the position of GpmiR156a and GpmiR166b,respectively
| GpmiR156a靶基因预测GpmiR156a target gene prediction | GpmiR166b靶基因预测GpmiR166b target gene prediction | |||
|---|---|---|---|---|
| 编号Serial No. | 名称Name | 编号Serial No. | 名称Name | |
| AT5G43270.3 | SPL2 | squamosa promoter binding protein-like 2 | AT1G30490.1 | PHV,ATHB9 | |
| AT2G33810.1 | SPL3 | squamosa promoter binding protein-like 3 | |||
| AT1G53160.1 | SPL4 | squamosa promoter binding protein-like 4 | AT1G52150.3 | ATHB-15,ATHB15,CNA | |
| AT3G57920.1 | SPL5 | squamosa promoter binding protein-like 5 | |||
| AT2G42200.1 | SPL9,AtSPL9 | squamosa promoter binding protein-like 9 | AT2G34710.1 | PHB,ATHB14,ATHB-14 | |
| AT1G27370.3 | SPL10 | squamosa promoter binding protein-like 10 | |||
| AT1G27360.4 | SPL11 | squamosa promoter-like 11 | AT4G32880.1 | ATHB-8,ATHB8,HB-8 | homeobox gene 8 | |
| AT5G50570.2 | SPL13A,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | |||
| AT5G50670.1 | SPL13B,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | AT5G60690.1 | REV,IFL | |
| AT3G57920.1 | SPL15 | squamosa promoter binding protein-like 15 | |||
表2 绞股蓝GpmiR156a和GpmiR166b的靶基因预测
Table 2 Prediction of G. pentaphyllu GpmiR156a and GpmiR166b target genes
| GpmiR156a靶基因预测GpmiR156a target gene prediction | GpmiR166b靶基因预测GpmiR166b target gene prediction | |||
|---|---|---|---|---|
| 编号Serial No. | 名称Name | 编号Serial No. | 名称Name | |
| AT5G43270.3 | SPL2 | squamosa promoter binding protein-like 2 | AT1G30490.1 | PHV,ATHB9 | |
| AT2G33810.1 | SPL3 | squamosa promoter binding protein-like 3 | |||
| AT1G53160.1 | SPL4 | squamosa promoter binding protein-like 4 | AT1G52150.3 | ATHB-15,ATHB15,CNA | |
| AT3G57920.1 | SPL5 | squamosa promoter binding protein-like 5 | |||
| AT2G42200.1 | SPL9,AtSPL9 | squamosa promoter binding protein-like 9 | AT2G34710.1 | PHB,ATHB14,ATHB-14 | |
| AT1G27370.3 | SPL10 | squamosa promoter binding protein-like 10 | |||
| AT1G27360.4 | SPL11 | squamosa promoter-like 11 | AT4G32880.1 | ATHB-8,ATHB8,HB-8 | homeobox gene 8 | |
| AT5G50570.2 | SPL13A,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | |||
| AT5G50670.1 | SPL13B,SPL13 | Squamosa promoter-binding protein-like(SBP domain)transcription factor family protein | AT5G60690.1 | REV,IFL | |
| AT3G57920.1 | SPL15 | squamosa promoter binding protein-like 15 | |||

图5 转基因植株的检测与表型分析 A:GpmiR156a和GpmiR166b转基因株系的部分PCR鉴定结果;M:marker;1:阴性对照;2:GpmiR156a的阳性对照;3-12:GpmiR156a;13:GpmiR166b的阳性对照;14-24:GpmiR166b;B:GpmiR156a和GpmiR166b转基因株系的GUS染色鉴定;1:野生型拟南芥;2-7:GpmiR156a转基因植株;8-13:GpmiR166b转基因植株;Bar=5 mm;C:Kan抗性筛选;a:GpmiR156a筛选培养基;b:GpmiR166b筛选培养基;红色箭头指示Kan抗性筛选出的转基因拟南芥;D:野生型与转基因拟南芥的表型;a,d:野生型拟南芥;b,e:GpmiR156a转基因拟南芥;c,f:GpmiR166b转基因拟南芥;Bar=1 cm
Fig. 5 Detection and phenotype analysis of positive transg-enic plants A:Partial PCR identification results of GpmiR156a and GpmiR166b transgenic lines. M:Marker. 1:Negative control. 2:Positive control of GpmiR156a. 3-12:GpmiR156a. 13:Positive control of GpmiR166b. 14-24:GpmiR166b. B:Identification of GpmiR156a and GpmiR166b transgenic lines by GUS staining. 1:Wild-type(WT)A. thaliana. 2-7:GpmiR156a transgenic plants. 8-13:GpmiR166b transgenic plants. Bar=5 mm. C:Kan resistance screening. a:GpmiR156a screening medium. b:GpmiR166b screening medium. Red arrows indicate the transgenic A. thaliana selected for Kan resistance. D:Phenotype of WT and transgenic A. thaliana. a,d:WT Arabidopsis thaliana. b,e:GpmiR156a transgenic A. thaliana. c,f:GpmiR166b transgenic A. thaliana. Bar=1 cm
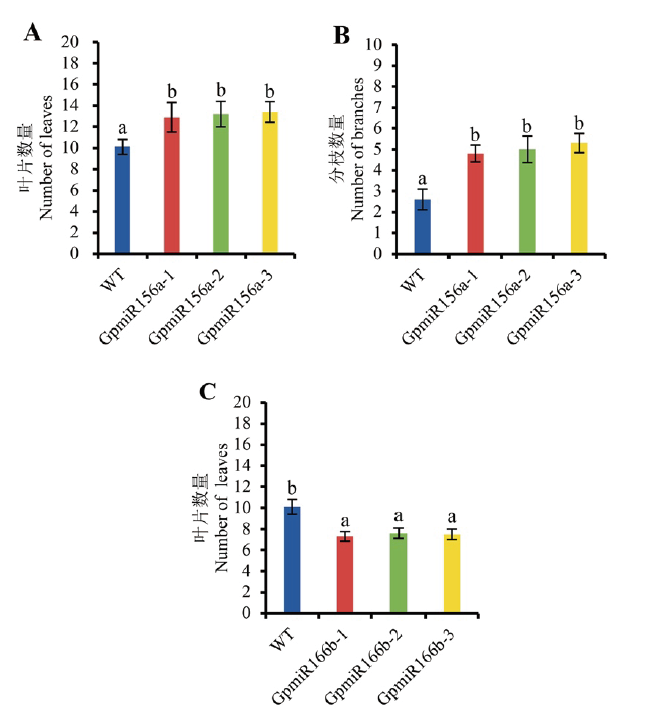
图6 转基因拟南芥与野生型拟南芥的叶片及分枝数量 A:过表达GpmiR156a拟南芥叶片数量;B:过表达GpmiR156a拟南芥分枝数量;C:过表达GpmiR166b拟南芥叶片数量;GpmiR156a-1,GpmiR156a-2,GpmiR156a-3为3个转基因株系;GpmiR166b-1,GpmiR166b-2,GpmiR166b-3为3个转基因株系;分枝数量指除主枝以外的数目。n=10。小写字母a,b表示野生型和转基因株系之间的差异显著性(P<0.05)。下同
Fig. 6 Number of rosette leaves and branches in transgenic A. thaliana and WT A:Number of rosette leaves in the transgenic A. thaliana overexpressing GpmiR156a. B:Number of branches in the transgenic A. thaliana overexpressing GpmiR156a. C:Number of rosette leaves in the transgenic A. thaliana overexpressing GpmiR166b. GpmiR156a-1,GpmiR156a-2,and GpmiR156a-3 are three separate lines of transgenic A. thaliana. GpmiR166b-1,GpmiR166b-2,and GpmiR166b-3 are three separate lines of transgenic A. thaliana. Number of branches refers to the branches other than the main branches. n=10. Lowercase letters a and b refer to the significant difference between wild type and transgenic plants. The same below
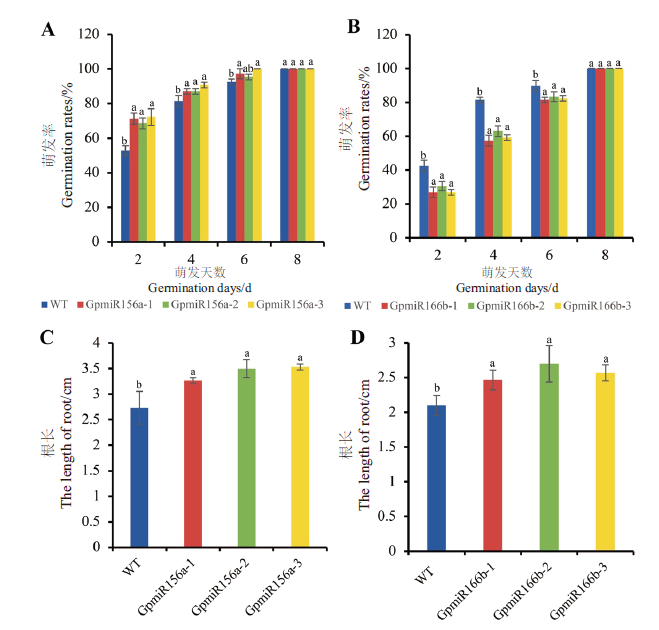
图7 过表达GpmiR156a、GpmiR166b拟南芥与野生型拟南芥种子的萌发与根长 A:过表达GpmiR156a拟南芥种子的萌发;B:过表达GpmiR166b拟南芥种子的萌发;C:过表达GpmiR156a拟南芥的根长;D:过表达GpmiR166b拟南芥的根长;GpmiR156a-1,GpmiR156a-2,GpmiR156a-3为3个转基因株系;GpmiR166b-1,GpmiR166b-2,GpmiR166b-3为3个转基因株系;A,B:n=36;C,D:n=5
Fig. 7 Seed germination and root length of overexpressing GpmiR156a and GpmiR166b and wild-type A. thal-iana A:Seed germination of transgenic Arabidopsis overexpressing GpmiR156a. B:Seed germination of transgenic Arabidopsis overexpressing GpmiR166b. C:Root length of transgenic Arabidopsisoverexpressing GpmiR156a. D:Root length of transgenic Arabidopsis overexpressing GpmiR166b. GpmiR156a-1,GpmiR156a-2,and GpmiR156a-3 are three separate lines of transgenic Arabidopsis. GpmiR166b-1,GpmiR166b-2,and GpmiR166b-3 are three separate lines of transgenic Arabidopsis. A,B:n=36;C,D:n=5
| [1] |
Razmovski-Naumovski V, Huang THW, Tran VH, et al. Chemistry and pharmacology of Gynostemma pentaphyllum[J]. Phytochem Rev, 2005, 4(2/3):197-219.
doi: 10.1007/s11101-005-3754-4 URL |
| [2] |
Yang Q, Liu SB, Han XN, et al. Integrated transcriptome and miRNA analysis uncovers molecular regulators of aerial stem-to-rhizome transition in the medical herb Gynostemma pentaphyllum[J]. BMC Genomics, 2019, 20(1):865.
doi: 10.1186/s12864-019-6250-8 pmid: 31730459 |
| [3] |
Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets[J]. Trends Plant Sci, 2008, 13(7):343-349.
doi: 10.1016/j.tplants.2008.03.009 URL |
| [4] |
Ma JY, Zhao P, Liu SB, et al. The control of developmental phase transitions by microRNAs and their targets in seed plants[J]. Int J Mol Sci, 2020, 21(6):1971.
doi: 10.3390/ijms21061971 URL |
| [5] |
Schwab R, Palatnik JF, Riester M, et al. Specific effects of microRNAs on the plant transcriptome[J]. Dev Cell, 2005, 8(4):517-527.
doi: 10.1016/j.devcel.2005.01.018 URL |
| [6] |
Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity[J]. Nat Genet, 2007, 39(8):1033-1037.
pmid: 17643101 |
| [7] | He J, Xu ML, Willmann MR, et al. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana[J]. PLoS Genet, 2018, 14(4):e1007337. |
| [8] |
Wang J, Gao XY, Li L, et al. Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice[J]. J Exp Bot, 2010, 61(6):1885-1895.
doi: 10.1093/jxb/erp378 URL |
| [9] |
Williams L, Grigg SP, Xie MT, et al. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes[J]. Development, 2005, 132(16):3657-3668.
pmid: 16033795 |
| [10] |
Jung JH, Park CM. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis[J]. Planta, 2007, 225(6):1327-1338.
doi: 10.1007/s00425-006-0439-1 URL |
| [11] | Wang JW, Park MY, Wang LJ, et al. miRNA control of vegetative phase change in trees[J]. PLoS Genet, 2011, 7(2):e1002012. |
| [12] | Xiong JS, Bai YB, Ma CJ, et al. Molecular cloning and characterization of SQUAMOSA-promoter binding protein-like gene FvSPL10 from woodland strawberry(Fragaria vesca)[J]. Plants(Basel), 2019, 8(9):342. |
| [13] |
Guo Q, Li L, Zhao K, et al. Genome-wide analysis of poplar SQUAMOSA-promoter-binding protein(SBP)family under salt stress[J]. Forests, 2021, 12(4):413.
doi: 10.3390/f12040413 URL |
| [14] | Xu ML, Hu TQ, Zhao JF, et al. Developmental functions of mir156-regulated Squamosa promoter binding protein-like(spl)genes in Arabidopsis thaliana[J]. PLoS Genet, 2016, 12(8):e1006263. |
| [15] |
Gao RM, Gruber MY, Amyot L, et al. SPL13 regulates shoot branching and flowering time in Medicago sativa[J]. Plant Mol Biol, 2018, 96(1/2):119-133.
doi: 10.1007/s11103-017-0683-8 URL |
| [16] |
Miao CB, Wang Z, Zhang L, et al. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice[J]. Nat Commun, 2019, 10(1):3822.
doi: 10.1038/s41467-019-11830-5 URL |
| [17] |
Zhang L, Ding H, Jiang HL, et al. Regulation of cadmium tolerance and accumulation by miR156 in Arabidopsis[J]. Chemosphere, 2020, 242:125168.
doi: 10.1016/j.chemosphere.2019.125168 URL |
| [18] |
Barrera-Rojas CH, Rocha GHB, Polverari L, et al. miR156-targeted SPL10 controls Arabidopsis root meristem activity and root-derived de novo shoot regeneration via cytokinin responses[J]. J Exp Bot, 2020, 71(3):934-950.
doi: 10.1093/jxb/erz475 pmid: 31642910 |
| [19] |
Willmann MR, Poethig RS. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis[J]. Development, 2011, 138(4):677-685.
doi: 10.1242/dev.057448 pmid: 21228003 |
| [20] | Tang XR, Bian SM, Tang MJ, et al. microRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis[J]. PLoS Genet, 2012, 8(11):e1003091. |
| [21] |
Li ZX, Zhang LF, Li WF, et al. MIR166a affects the germination of somatic embryos in larixleptolepis by modulating IAA biosynthesis and signaling genes[J]. J Plant Growth Regul, 2017, 36(4):889-896.
doi: 10.1007/s00344-017-9693-7 URL |
| [22] |
Barik S, SarkarDas S, Singh A, et al. Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species[J]. Genomics, 2014, 103(1):114-121.
doi: 10.1016/j.ygeno.2013.11.004 URL |
| [23] |
Chaves I, Lin YC, Pinto-Ricardo C, et al. miRNA profiling in leaf and cork tissues of Quercus suber reveals novel miRNAs and tissue-specific expression patterns[J]. Tree Genet Genomes, 2014, 10(3):721-737.
doi: 10.1007/s11295-014-0717-1 URL |
| [24] |
Li ZX, Qi LW. Over-expression of LaMIR166a promotes organs development in Nicotiana benthamiana[J]. Russ J Plant Physiol, 2019, 66(5):718-724.
doi: 10.1134/S1021443719050133 URL |
| [25] | Gautam V, Singh A, Yadav S, et al. Conserved LBL1-ta-siRNA and miR165/166-RLD1/2 modules regulate root development in maize[J]. Development, 2021, 148(1):dev190033. |
| [1] | 胡明月, 杨宇, 郭仰东, 张喜春. 低温胁迫下番茄SlMYB96的功能分析[J]. 生物技术通报, 2023, 39(4): 236-245. |
| [2] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [3] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [4] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| [5] | 陈浩婷, 张玉静, 刘洁, 代泽敏, 刘伟, 石玉, 张毅, 李天来. 低磷胁迫下番茄转录因子WRKY6功能分析[J]. 生物技术通报, 2023, 39(10): 136-147. |
| [6] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [7] | 陈佳敏, 刘永杰, 马锦绣, 李丹, 公杰, 赵昌平, 耿洪伟, 高世庆. 小麦组蛋白甲基化酶在杂交种中干旱胁迫表达模式分析[J]. 生物技术通报, 2022, 38(7): 51-61. |
| [8] | 王楠, 张瑞, 潘阳阳, 何翃宏, 王靖雷, 崔燕, 余四九. 牦牛TGF-β1基因克隆及在雌性生殖系统主要器官中的表达定位[J]. 生物技术通报, 2022, 38(6): 279-290. |
| [9] | 李宇航, 王兴平, 杨箭, 罗仍卓么, 任倩倩, 魏大为, 马云. miR-665在奶牛乳腺上皮细胞炎症中的表达及功能分析[J]. 生物技术通报, 2022, 38(5): 159-168. |
| [10] | 李洋, 张晓天, 朴静子, 周如军, 李自博, 关海雯. 花生疮痂病菌蓝光受体EaWC 1基因克隆及生物信息学分析[J]. 生物技术通报, 2022, 38(5): 93-99. |
| [11] | 张琳, 魏祯祯, 宋程威, 郭丽丽, 郭琪, 侯小改, 王华芳. ‘凤丹’牡丹PoFD基因克隆及表达分析[J]. 生物技术通报, 2022, 38(11): 104-111. |
| [12] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| [13] | 郑青波, 叶娜, 张哓兰, 包鹏甲, 王福彬, 任稳稳, 廖月姣, 阎萍, 潘和平. 天祝白牦牛退行期毛囊细胞亚群鉴定以及特征基因生物信息学分析[J]. 生物技术通报, 2022, 38(10): 262-272. |
| [14] | 范亚朋, 芮存, 张悦新, 陈修贵, 陆许可, 王帅, 张红, 徐楠, 王晶, 陈超, 叶武威. 陆地棉耐碱基因GHZAT12的克隆、表达及生物信息学分析[J]. 生物技术通报, 2021, 37(8): 121-130. |
| [15] | 杜振伟, 朱帅鹏, 马向飞, 李东华, 孙桂荣. 鸡CEBPA基因CDS区克隆、表达及生物信息学分析[J]. 生物技术通报, 2021, 37(8): 203-212. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||