








生物技术通报 ›› 2023, Vol. 39 ›› Issue (11): 308-317.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0096
张玉娟1( ), 黎冬华2, 宫慧慧1, 崔新晓1, 高春华1, 张秀荣1, 游均2(
), 黎冬华2, 宫慧慧1, 崔新晓1, 高春华1, 张秀荣1, 游均2( ), 赵军胜1(
), 赵军胜1( )
)
收稿日期:2023-02-08
出版日期:2023-11-26
发布日期:2023-12-20
通讯作者:
赵军胜,男,博士,研究员,研究方向:经济作物遗传育种;E-mail: zhaojunshengsd@163.com;作者简介:张玉娟,女,博士,助理研究员,研究方向:植物逆境分子生物学;E-mail: 723949246@qq.com
基金资助:
ZHANG Yu-juan1( ), LI Dong-hua2, GONG Hui-hui1, CUI Xin-xiao1, GAO Chun-hua1, ZHANG Xiu-rong1, YOU Jun2(
), LI Dong-hua2, GONG Hui-hui1, CUI Xin-xiao1, GAO Chun-hua1, ZHANG Xiu-rong1, YOU Jun2( ), ZHAO Jun-sheng1(
), ZHAO Jun-sheng1( )
)
Received:2023-02-08
Published:2023-11-26
Online:2023-12-20
摘要:
NAC转录因子在植物生长发育及盐胁迫响应过程中具有重要的调控作用。芝麻SiNAC77参与盐胁迫响应过程。克隆芝麻SiNAC77并分析其耐盐功能,为芝麻耐盐育种提供分子基础和基因资源。克隆SiNAC77的全长CDS序列,利用生物信息学软件对其基因序列和氨基酸序列特征进行分析。构建SiNAC77过表达载体,利用农杆菌介导法转化拟南芥。对过表达株系的耐盐表型和生理生化指标进行分析。结果表明,成功克隆了长度为1 008 bp的SiNAC77 CDS序列,编码335个氨基酸,相对分子量为135.93 kD,预测等电点为4.91,含有33个潜在的磷酸化位点;启动子区域含有MBS、STRE、ARE、ABRE和TCA等多个非生物胁迫及激素响应元件。成功构建了SiNAC77过表达载体,并转化拟南芥,获得12个独立的转基因阳性株系。在盐胁迫下,与野生型拟南芥相比,过表达株系的种子发芽率、初生根长度、鲜重均显著提高,转基因株系中Na+/K+和相对MDA含量显著降低,相对SOD和POD抗氧化酶活性则显著增强。过表达芝麻SiNAC77可以增强抗氧化酶活性,减少离子毒害和氧化损伤,提高转基因植株的耐盐性。
张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317.
ZHANG Yu-juan, LI Dong-hua, GONG Hui-hui, CUI Xin-xiao, GAO Chun-hua, ZHANG Xiu-rong, YOU Jun, ZHAO Jun-sheng. Cloning and Salt-tolerance Analysis of NAC Transcription Factor SiNAC77 from Sesamum indicum L.[J]. Biotechnology Bulletin, 2023, 39(11): 308-317.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| NAC77-BamH I-F | tgtctagactcgagggatccATGGAACAGTTGGCCGGTTTTG | 基因克隆Gene clone |
| NAC77-BamH I-R | agcctgcagccatggggatccTCAGTAGTTCCACAAGCAGTC | |
| 35s-F | GACGCACAATCCCACTATCC | 转基因植株检测Transgenic plant detection |
| NAC77-R | TCAGTAGTTCCACAAGCAGTC | |
| Actin-F | CGGCGAGTCATCGTCATCAATGT | 拟南芥内参引物Internal primer of Arabidopsis thaliana |
| Actin-R | GTTCGGGATTACGGTGGTGGGA | |
| NAC77-Fq | CTCCACTCACGGACTTGTCA | 荧光定量PCR引物Fluorescent quantitative PCR primer |
| NAC77-Rq | AGGAAAGGAGGGTTGGATTG |
表1 本研究所用引物序列
Table 1 Primer sequence used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| NAC77-BamH I-F | tgtctagactcgagggatccATGGAACAGTTGGCCGGTTTTG | 基因克隆Gene clone |
| NAC77-BamH I-R | agcctgcagccatggggatccTCAGTAGTTCCACAAGCAGTC | |
| 35s-F | GACGCACAATCCCACTATCC | 转基因植株检测Transgenic plant detection |
| NAC77-R | TCAGTAGTTCCACAAGCAGTC | |
| Actin-F | CGGCGAGTCATCGTCATCAATGT | 拟南芥内参引物Internal primer of Arabidopsis thaliana |
| Actin-R | GTTCGGGATTACGGTGGTGGGA | |
| NAC77-Fq | CTCCACTCACGGACTTGTCA | 荧光定量PCR引物Fluorescent quantitative PCR primer |
| NAC77-Rq | AGGAAAGGAGGGTTGGATTG |

图4 转基因拟南芥植株的鉴定 A:卡那霉素筛选T1转基因苗;B:T1转基因植株的PCR鉴定,1-12为转基因植株,“-”为纯水空白对照,“+”为质粒对照,WT为野生型植株对照,M为DL2000 marker;C:T2转基因拟南芥中SiNAC77的表达量检测,OE-1、OE-2和OE-3分别来自T1“4、5、8”转基因拟南芥家系。*表示在P<0.05水平上差异显著,**表示在P<0.01水平上差异显著。下同
Fig. 4 Identification of transgenic Arabidopsis plants A: Identification of T1 transgenic Arabidopsis plants by kanamycin. B: Identification of T1 transgenic Arabidopsis plants, 1-12 indicate transgenic lines, “-” indicates pure water blank control, “+” indicates plasmid control, WT indicates wild type of Arabidopsis plants control, and M indicates DL2000 marker. C: Expression of SiNAC77in transgenic Arabidopsis. OE-1, OE-2, and OE-3 were obtained from T1(4, 5 and 8)transgenic plants. * indicate significant difference at P<0.05 level; **indicates significant difference at P<0.01 level. The same below
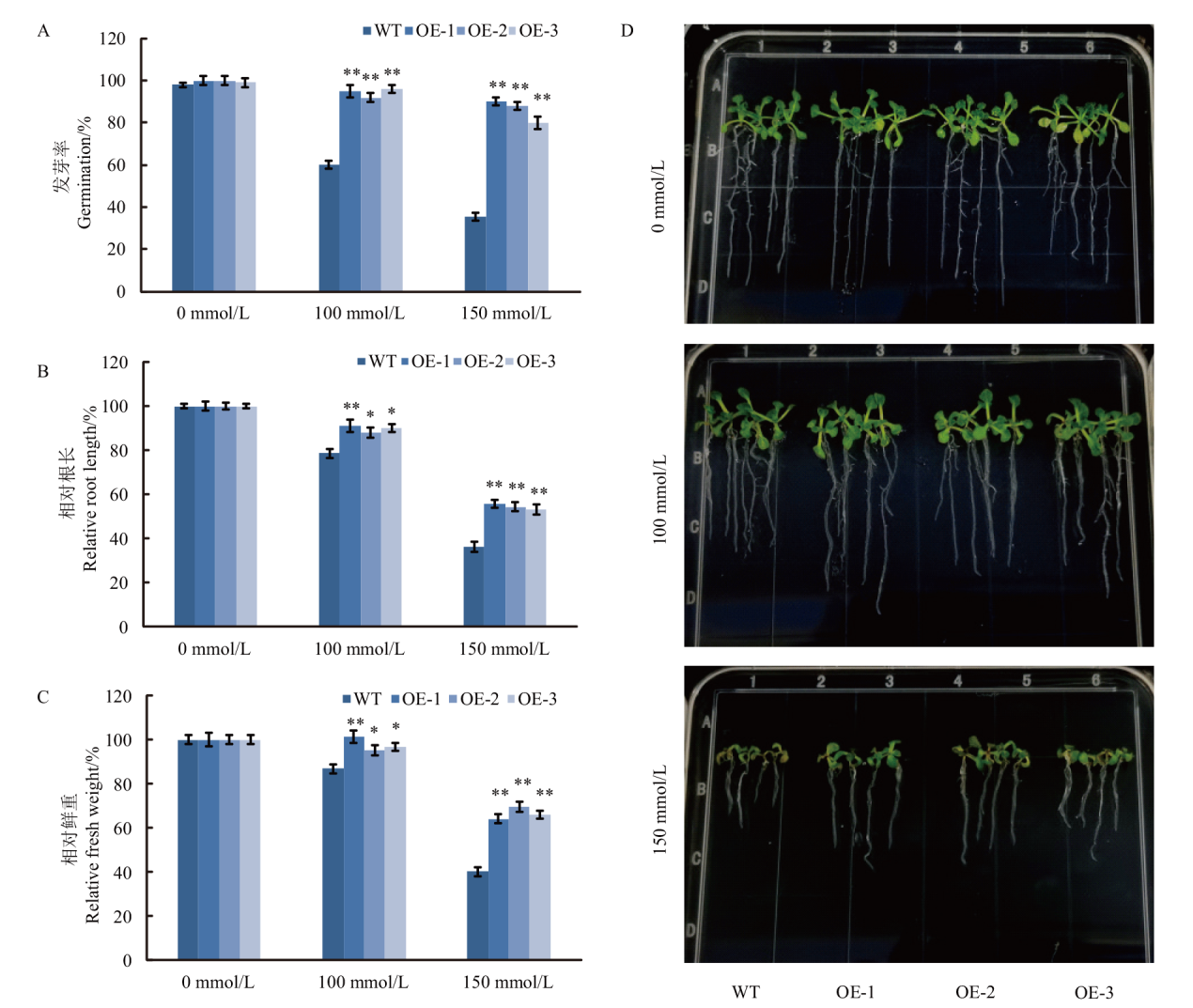
图5 转基因拟南芥的耐盐性鉴定 A:盐胁迫对转基因拟南芥种子发芽率的影响;B:盐胁迫对转基因拟南芥初生根根长的影响;C:盐胁迫对转基因拟南芥生物积累量的影响;D:盐胁迫下野生型和转基因拟南芥植株的表型
Fig. 5 Resistance characteristics of transgenic Arabidopsis plants to salt stress A: Seed germination of transgenic Arabidopsis under salt stress. B: The elongation of the primary root of transgenic Arabidopsis under salt stress. C: Fresh weight of transgenic Arabidopsis under salt stress. D: Phenotypes of wild type and transgenic Arabidopsis plants under salt stress
| [1] |
Namiki M. Nutraceutical functions of sesame: a review[J]. Crit Rev Food Sci Nutr, 2007, 47(7): 651-673.
pmid: 17943496 |
| [2] |
Murata J, Ono E, Yoroizuka S, et al. Oxidative rearrangement of(+)-sesamin by CYP92B14 co-generates twin dietary lignans in sesame[J]. Nat Commun, 2017, 8(1): 2155.
doi: 10.1038/s41467-017-02053-7 pmid: 29255253 |
| [3] |
Harikumar KB, Sung B, Tharakan ST, et al. Sesamin manifests chemopreventive effects through the suppression of NF-kappa B-regulated cell survival, proliferation, invasion, and angiogenic gene products[J]. Mol Cancer Res, 2010, 8(5): 751-761.
doi: 10.1158/1541-7786.MCR-09-0565 pmid: 20460401 |
| [4] |
Khan MM, Ishrat T, Ahmad A, et al. Sesamin attenuates behavioral, biochemical and histological alterations induced by reversible middle cerebral artery occlusion in the rats[J]. Chem Biol Interact, 2010, 183(1): 255-263.
doi: 10.1016/j.cbi.2009.10.003 URL |
| [5] |
Ernst HA, Olsen AN, Larsen S, et al. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors[J]. EMBO Rep, 2004, 5(3): 297-303.
doi: 10.1038/sj.embor.7400093 pmid: 15083810 |
| [6] |
Puranik S, Sahu PP, Srivastava PS, et al. NAC proteins: regulation and role in stress tolerance[J]. Trends Plant Sci, 2012, 17(6): 369-381.
doi: 10.1016/j.tplants.2012.02.004 pmid: 22445067 |
| [7] |
Ismail AM, Horie T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance[J]. Annu Rev Plant Biol, 2017, 68: 405-434.
doi: 10.1146/annurev-arplant-042916-040936 pmid: 28226230 |
| [8] |
van Zelm E, Zhang YX, Testerink C. Salt tolerance mechanisms of plants[J]. Annu Rev Plant Biol, 2020, 71: 403-433.
doi: 10.1146/annurev-arplant-050718-100005 pmid: 32167791 |
| [9] |
Wang YP, Cui Y, Liu B, et al. Lilium pumilum stress-responsive NAC transcription factor LpNAC17 enhances salt stress tolerance in tobacco[J]. Front Plant Sci, 2022, 13: 993841.
doi: 10.3389/fpls.2022.993841 URL |
| [10] |
Yuan CL, Li CJ, Lu XD, et al. Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut[J]. BMC Plant Biol, 2020, 20(1): 454.
doi: 10.1186/s12870-020-02678-9 pmid: 33008287 |
| [11] |
Li M, Chen R, Jiang QY, et al. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean[J]. Plant Mol Biol, 2021, 105(3): 333-345.
doi: 10.1007/s11103-020-01091-y pmid: 33155154 |
| [12] |
Fang YJ, You J, Xie KB, et al. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice[J]. Mol Genet Genomics, 2008, 280(6): 547-563.
doi: 10.1007/s00438-008-0386-6 pmid: 18813954 |
| [13] |
Hu HH, Dai MQ, Yao JL, et al. Overexpressing a NAM, ATAF, and CUC(NAC)transcription factor enhances drought resistance and salt tolerance in rice[J]. Proc Natl Acad Sci USA, 2006, 103(35): 12987-12992.
doi: 10.1073/pnas.0604882103 URL |
| [14] |
Zhang X, Long Y, Chen XX, et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice[J]. BMC Plant Biol, 2021, 21(1): 546.
doi: 10.1186/s12870-021-03333-7 pmid: 34800972 |
| [15] |
Takasaki H, Maruyama K, Kidokoro S, et al. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice[J]. Mol Genet Genomics, 2010, 284(3): 173-183.
doi: 10.1007/s00438-010-0557-0 pmid: 20632034 |
| [16] |
Zheng XN, Chen B, Lu GJ, et al. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance[J]. Biochem Biophys Res Commun, 2009, 379(4): 985-989.
doi: 10.1016/j.bbrc.2008.12.163 URL |
| [17] |
Wang Q, Guo C, Li ZY, et al. Potato NAC transcription factor StNAC053 enhances salt and drought tolerance in transgenic Arabidopsis[J]. Int J Mol Sci, 2021, 22(5): 2568.
doi: 10.3390/ijms22052568 URL |
| [18] |
Bekele A, Besufekad Y, Adugna S, et al. Screening of selected accessions of Ethiopian sesame(Sesame indicum L.) for salt tolerance[J]. Biocatal Agric Biotechnol, 2017, 9: 82-94.
doi: 10.1016/j.bcab.2016.11.009 URL |
| [19] |
Radhakrishnan R, Lee IJ. Penicillium-sesame interactions: a remedy for mitigating high salinity stress effects on primary and defense metabolites in plants[J]. Environ Exp Bot, 2015, 116: 47-60.
doi: 10.1016/j.envexpbot.2015.03.008 URL |
| [20] |
Li DH, Dossa K, Zhang YX, et al. GWAS uncovers differential genetic bases for drought and salt tolerances in sesame at the germination stage[J]. Genes, 2018, 9(2): 87.
doi: 10.3390/genes9020087 URL |
| [21] |
Zhang YJ, Li DH, Zhou R, et al. Transcriptome and metabolome analyses of two contrasting sesame genotypes reveal the crucial biological pathways involved in rapid adaptive response to salt stress[J]. BMC Plant Biol, 2019, 19(1): 66.
doi: 10.1186/s12870-019-1665-6 pmid: 30744558 |
| [22] |
Zhang YJ, Wei MY, Liu AL, et al. Comparative proteomic analysis of two sesame genotypes with contrasting salinity tolerance in response to salt stress[J]. J Proteomics, 2019, 201: 73-83.
doi: S1874-3919(19)30130-7 pmid: 31009803 |
| [23] |
Zhang YJ, Li DH, Zhou R, et al. A collection of transcriptomic and proteomic datasets from sesame in response to salt stress[J]. Data Brief, 2020, 32: 106096.
doi: 10.1016/j.dib.2020.106096 URL |
| [24] |
Dossa K, Mmadi MA, Zhou R, et al. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis[J]. AoB Plants, 2019, 12(1): plz081.
doi: 10.1093/aobpla/plz081 URL |
| [25] |
Zhang YJ, Li DH, Wang YY, et al. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum[J]. PLoS One, 2018, 13(6): e0199262.
doi: 10.1371/journal.pone.0199262 URL |
| [26] | 郭安源, 朱其慧, 陈新, 等. GSDS: 基因结构显示系统[J]. 遗传, 2007, 29(8): 1023-1026. |
| Guo AY, Zhu QH, Chen X, et al. GSDS: a gene structure display server[J]. Hereditas, 2007, 29(8): 1023-1026. | |
| [27] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [28] |
Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server[J]. Methods Mol Biol, 1999, 112: 531-552.
pmid: 10027275 |
| [29] |
Jin JP, Tian F, Yang DC, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants[J]. Nucleic Acids Res, 2017, 45(D1): D1040-D1045.
doi: 10.1093/nar/gkw982 URL |
| [30] |
Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms[J]. Nat Protoc, 2008, 3(2): 153-162.
doi: 10.1038/nprot.2007.494 |
| [31] |
Blom N, Sicheritz-Pontén T, Gupta R, et al. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence[J]. Proteomics, 2004, 4(6): 1633-1649.
doi: 10.1002/pmic.200300771 pmid: 15174133 |
| [32] |
刘爱丽, 魏梦园, 黎冬华, 等. 芝麻肌醇半乳糖苷合成酶基因SiGolS6的克隆及功能分析[J]. 中国农业科学, 2020, 53(17): 3432-3442.
doi: 10.3864/j.issn.0578-1752.2020.17.002 |
| Liu AL, Wei MY, Li DH, et al. Cloning and function analysis of sesame galactinol synthase gene SiGolS6 in Arabidopsis[J]. Sci Agric Sin, 2020, 53(17): 3432-3442. | |
| [33] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [34] |
Ghaffari A, Gharechahi J, Nakhoda B, et al. Physiology and proteome responses of two contrasting rice mutants and their wild type parent under salt stress conditions at the vegetative stage[J]. J Plant Physiol, 2014, 171(1): 31-44.
doi: 10.1016/j.jplph.2013.07.014 URL |
| [35] |
Badis G, Berger MF, Philippakis AA, et al. Diversity and complexity in DNA recognition by transcription factors[J]. Science, 2009, 324(5935): 1720-1723.
doi: 10.1126/science.1162327 pmid: 19443739 |
| [36] |
Cheong YH, Kim KN, Pandey GK, et al. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis[J]. Plant Cell, 2003, 15(8): 1833-1845.
pmid: 12897256 |
| [37] |
Shi J, Kim KN, Ritz O, et al. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis[J]. Plant Cell, 1999, 11(12): 2393-2405.
doi: 10.1105/tpc.11.12.2393 pmid: 10590166 |
| [38] |
He XJ, Mu RL, Cao WH, et al. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development[J]. Plant J, 2005, 44(6): 903-916.
doi: 10.1111/tpj.2005.44.issue-6 URL |
| [39] |
Sun Y, Song KQ, Guo MM, et al. A NAC transcription factor from ‘sea rice 86’ enhances salt tolerance by promoting hydrogen sulfide production in rice seedlings[J]. Int J Mol Sci, 2022, 23(12): 6435.
doi: 10.3390/ijms23126435 URL |
| [40] |
Jiang DG, Zhou LY, Chen WT, et al. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in rice via ABA-mediated pathways[J]. Rice, 2019, 12(1): 76.
doi: 10.1186/s12284-019-0334-6 pmid: 31637532 |
| [41] |
Ma J, Wang LY, Dai JX, et al. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana[J]. BMC Plant Biol, 2021, 21(1): 11.
doi: 10.1186/s12870-020-02764-y |
| [42] |
Yarra R, Wei W. The NAC-type transcription factor GmNAC20 improves cold, salinity tolerance, and lateral root formation in transgenic rice plants[J]. Funct Integr Genomics, 2021, 21(3/4): 473-487.
doi: 10.1007/s10142-021-00790-z |
| [43] | 杨勇, 田新权, 刘会利, 等. 陆地棉NAC转录因子基因GhNAC6的克隆、表达和耐盐性分析[J]. 棉花学报, 2017, 29(2): 138-146. |
| Yang Y, Tian XQ, Liu HL, et al. Cloning, expression and salt-tolerance analysis of the NAC transcription factor gene GhNAC6 in upland cotton[J]. Cotton Sci, 2017, 29(2): 138-146. | |
| [44] |
Roy SJ, Negrão S, Tester M. Salt resistant crop plants[J]. Curr Opin Biotechnol, 2014, 26: 115-124.
doi: 10.1016/j.copbio.2013.12.004 URL |
| [45] |
Deinlein U, Stephan AB, Horie T, et al. Plant salt-tolerance mechanisms[J]. Trends Plant Sci, 2014, 19(6): 371-379.
doi: 10.1016/j.tplants.2014.02.001 pmid: 24630845 |
| [46] |
You J, Chan ZL. ROS regulation during abiotic stress responses in crop plants[J]. Front Plant Sci, 2015, 6: 1092.
doi: 10.3389/fpls.2015.01092 pmid: 26697045 |
| [47] |
Xu ZY, Gongbuzhaxi, Wang CY, et al. Wheat NAC transcription factor TaNAC29 is involved in response to salt stress[J]. Plant Physiol Biochem, 2015, 96: 356-363.
doi: 10.1016/j.plaphy.2015.08.013 URL |
| [1] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [2] | 魏茜雅, 秦中维, 梁腊梅, 林欣琪, 李映志. 褪黑素种子引发处理提高朝天椒耐盐性的作用机制[J]. 生物技术通报, 2023, 39(7): 160-172. |
| [3] | 胡明月, 杨宇, 郭仰东, 张喜春. 低温胁迫下番茄SlMYB96的功能分析[J]. 生物技术通报, 2023, 39(4): 236-245. |
| [4] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [5] | 杜清洁, 周璐瑶, 杨思震, 张嘉欣, 陈春林, 李娟起, 李猛, 赵士文, 肖怀娟, 王吉庆. 过表达CaCP1提高转基因烟草对盐胁迫的敏感性[J]. 生物技术通报, 2023, 39(2): 172-182. |
| [6] | 叶红, 王玉昆. 植物PRR免疫受体功能研究进展[J]. 生物技术通报, 2023, 39(12): 1-15. |
| [7] | 汪明滔, 刘建伟, 赵春钊. 植物调控盐胁迫下细胞壁完整性的分子机制[J]. 生物技术通报, 2023, 39(11): 18-27. |
| [8] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| [9] | 陈浩婷, 张玉静, 刘洁, 代泽敏, 刘伟, 石玉, 张毅, 李天来. 低磷胁迫下番茄转录因子WRKY6功能分析[J]. 生物技术通报, 2023, 39(10): 136-147. |
| [10] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| [11] | 陈光, 李佳, 杜瑞英, 王旭. 水稻盐敏感突变体ss2的鉴定与基因功能分析[J]. 生物技术通报, 2022, 38(9): 158-166. |
| [12] | 于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193. |
| [13] | 张斌, 杨昕霞. 水稻响应盐胁迫关键转录因子的鉴定[J]. 生物技术通报, 2022, 38(3): 9-15. |
| [14] | 张业猛, 朱丽丽, 陈志国. 藜麦NHX基因家族鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2022, 38(12): 184-193. |
| [15] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||