








生物技术通报 ›› 2022, Vol. 38 ›› Issue (8): 188-197.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1492
史亚楠1( ), 王德培1,3, 王一川1, 周昊4, 薛鲜丽1,2(
), 王德培1,3, 王一川1, 周昊4, 薛鲜丽1,2( )
)
收稿日期:2021-12-02
出版日期:2022-08-26
发布日期:2022-09-14
作者简介:史亚楠,女,硕士研究生,研究方向:丝状真菌遗传改造;E-mail: 基金资助:
SHI Ya-nan1( ), WANG De-pei1,3, WANG Yi-chuan1, ZHOU Hao4, XUE Xian-li1,2(
), WANG De-pei1,3, WANG Yi-chuan1, ZHOU Hao4, XUE Xian-li1,2( )
)
Received:2021-12-02
Published:2022-08-26
Online:2022-09-14
摘要:
Msn2作为C2H2型锌指结构转录激活因子,参与调控真菌生长速率、分生孢子发育及多种应激反应等生物学过程。本论文以产曲酸米曲霉3.042为出发菌株,构建pyrG营养缺陷型菌株,通过msn2基因的上下游序列为同源重组臂和回补pyrG基因为筛选标记,最终获得一株敲除msn2且回补pyrG基因的转化子g-5号菌株。较原始菌株米曲霉3.042相比,g-5菌株生长受到抑制且孢子生成量显著降低60%,在含10%及以上葡萄糖环境中不产孢,但可耐受30 mmol/L H2O2。g-5菌株摇瓶发酵曲酸产量达到21.55 g/L,较米曲霉3.042的11.86 g/L,提升了81%。经RT-qPCR分析,与米曲霉3.042相比,g-5菌株的msn2基因不表达;在发酵6 d时米曲霉调节曲酸合成相关基因kojR和LaeA基因表达量分别提高4.13倍和21倍;g-5菌株菌丝生长及产孢相关基因brlA与tps1表达量较3.042下调63倍和128倍,相反ageB基因表达量大幅上调59倍。
史亚楠, 王德培, 王一川, 周昊, 薛鲜丽. 敲除msn2对米曲霉生长和发酵产曲酸的影响[J]. 生物技术通报, 2022, 38(8): 188-197.
SHI Ya-nan, WANG De-pei, WANG Yi-chuan, ZHOU Hao, XUE Xian-li. Effects of msn2 Knock-out on the Growth and Kojic Acid Production of Aspergillus oryzae[J]. Biotechnology Bulletin, 2022, 38(8): 188-197.
| 目的基因Target gene | 引物序列Sequence of primer(5'-3') |
|---|---|
| brlA | TTATCACAAGGCGGAGAAGG |
| CGAACAGTCAGTATGGGTTGC | |
| tps1 | GAATACGGGACTGGCCCT |
| GGGCTCGAAACATATCTGTGA | |
| ageB | CACCATCTCCAACCACTCG |
| CAGCGTAGGTGGGAGACC | |
| LaeA | TCCATTCATAGCAAGAGTTCTGAA |
| TTGCTCCTGCTCATCGC | |
| kojR | CCGCAGACGTTAGCGTAAAT |
| GCGATCGGCCATATTCATAC | |
| his2A | GCCTGGTGGAAAGGGAAAG |
| CTTTGCGCTGTGGGACTT | |
| msn2 | AAGCGAGGCTGTGACTTTG |
| AGGCGCTGTCTCTTTTCG |
表1 RT-qPCR 的引物
Table 1 RT-qPCR primers
| 目的基因Target gene | 引物序列Sequence of primer(5'-3') |
|---|---|
| brlA | TTATCACAAGGCGGAGAAGG |
| CGAACAGTCAGTATGGGTTGC | |
| tps1 | GAATACGGGACTGGCCCT |
| GGGCTCGAAACATATCTGTGA | |
| ageB | CACCATCTCCAACCACTCG |
| CAGCGTAGGTGGGAGACC | |
| LaeA | TCCATTCATAGCAAGAGTTCTGAA |
| TTGCTCCTGCTCATCGC | |
| kojR | CCGCAGACGTTAGCGTAAAT |
| GCGATCGGCCATATTCATAC | |
| his2A | GCCTGGTGGAAAGGGAAAG |
| CTTTGCGCTGTGGGACTT | |
| msn2 | AAGCGAGGCTGTGACTTTG |
| AGGCGCTGTCTCTTTTCG |

图1 质粒pk1的构建及△msn2菌株的筛选 A:pk1质粒示意图;B:msn2敲除示意图;C:转化子的平板筛选;D:△msn2菌株的PCR验证。M:DL5000;1:以3.042基因组为模板作为阴性对照(2 965 bp);2:质粒pk1为模板作为阳性对照(3 706 bp);3-5:转化子,其中3为质粒随机插入,4为质粒未插入,5为阳性转化子,6为以5号转化子基因组为PCR扩增msn2L上游到-pyrG中间(2 602 bp)
Fig.1 Construction of plasmid pk1 and screening of △msn2 strain A:pk1 plasmid diagram. B:msn2 knock-out diagram. C:Screening of transformants on plate. D:PCR verification of △msn2 strains. M:DL5000;1:negative control with genome template in 3.042(2 965 bp);2:positive control with pk1 template(3 706 bp);3-5:transformants,3 refers to that pk1 was randomly inserted in it,4 refers to that pk1 was not inserted in it,5 is positive transformant,and 6 refers to msn2L up stream-pyrG middle(2 602 bp)was validation by PCR with transformant 5 genome as template
| 菌株Strain | 24 h | 36 h | 48 h |
|---|---|---|---|
| 3.042 |  |  |  |
| g-5 |  |  |  |
表2 米曲霉3.042和g-5菌株生长形态观察
Table 2 Growth morphology of Aspergillus oryzae 3.042 and g-5 strain
| 菌株Strain | 24 h | 36 h | 48 h |
|---|---|---|---|
| 3.042 |  |  |  |
| g-5 |  |  |  |
| 时间 Time/h | 菌株 Strain | 0 mmol·L-1 | 15 mmol·L-1 | 25 mmol·L-1 | 30 mmol·L-1 |
|---|---|---|---|---|---|
| 36 | 3.042 |  |  |  |  |
| g-5 |  |  |  |  | |
| 48 | 3.042 |  |  |  |  |
| g-5 |  |  |  |  |
表3 米曲霉3.042和g-5菌株的H2O2耐受性
Table 3 H2O2 tolerance of A. oryzae 3.042 and g-5 strain
| 时间 Time/h | 菌株 Strain | 0 mmol·L-1 | 15 mmol·L-1 | 25 mmol·L-1 | 30 mmol·L-1 |
|---|---|---|---|---|---|
| 36 | 3.042 |  |  |  |  |
| g-5 |  |  |  |  | |
| 48 | 3.042 |  |  |  |  |
| g-5 |  |  |  |  |
| 时间 Time/h | 菌株Strain | 2% | 10% | 15% |
|---|---|---|---|---|
| 36 | 3.042 |  |  |  |
| g-5 |  |  |  | |
| 48 | 3.042 |  |  |  |
| g-5 |  |  |  |
表4 米曲霉3.042和g-5菌株高糖耐受性
Table 4 High sugar tolerance of A. oryzae 3.042 and g-5 strains
| 时间 Time/h | 菌株Strain | 2% | 10% | 15% |
|---|---|---|---|---|
| 36 | 3.042 |  |  |  |
| g-5 |  |  |  | |
| 48 | 3.042 |  |  |  |
| g-5 |  |  |  |
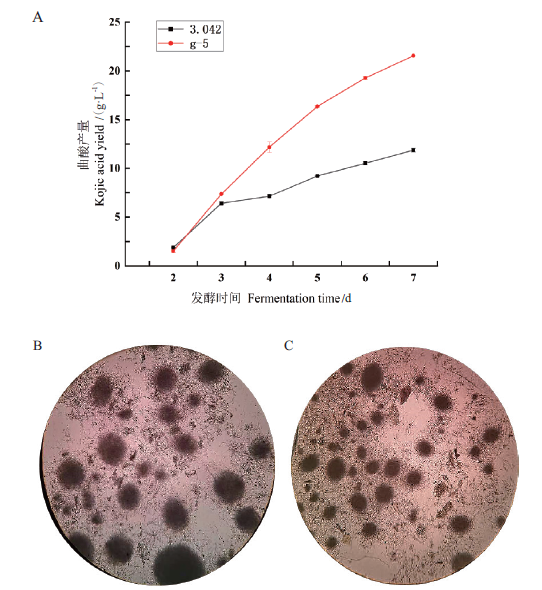
图2 米曲霉3.042和g-5号菌株发酵产曲酸分析及菌丝球形态观察 A:米曲霉3.042和g-5号菌株发酵产曲酸分析;B:3.042发酵7 d菌丝球形态;C:g-5菌株发酵7d菌丝球形态
Fig. 2 Fermentation results of A. oryzae 3.042 and strain g-5 and observation of mycelial pellet morphology A:Kojic acid yield of A. oryzae 3.042 and g-5 strain. B:Mycelial pellet morphology of A. oryzae 3.042 in at 7 d in fermentation. C:Mycelial pellet morphology of g-5 strain at 7 d of fermentation
| [1] | 易灿, 刘进兵. 曲酸衍生物的合成研究进展[J]. 山东化工, 2021, 50(5):74-78. |
| Yi C, Liu JB. Research progress in synthesis of kojic acid derivatives[J]. Shandong Chem Ind, 2021, 50(5):74-78. | |
| [2] |
Fan JX, Zhang Z, Long CN, et al. Identification and functional characterization of glycerol dehydrogenase reveal the role in kojic acid synthesis in Aspergillus oryzae[J]. World J Microbiol Biotechnol, 2020, 36(9):136.
doi: 10.1007/s11274-020-02912-4 URL |
| [3] | 罗丽娟, 王刚, 万玉军, 等. 曲酸美白护肤霜的研制[J]. 食品与发酵科技, 2019, 55(3):73-75, 94. |
| Luo LJ, Wang G, Wan YJ, et al. Development of kojic acid whitening cream[J]. Food Ferment Sci Technol, 2019, 55(3):73-75, 94. | |
| [4] | 张理珉, 程立忠, 陆和生. 三氯化铁比色法测定曲酸含量方法的改进[J]. 生物技术, 2000, 10(3):49-. |
| Zhang LM, Cheng LZ, Lu HS. Improvement of ferric chloride colorimetric method for the determination of kojic acid[J]. Biotechnology, 2000, 10(3):49-. | |
| [5] |
das Neves PAPFG, Lobato CC, Ferreira LR, et al. Molecular modification approach on kojic acid derivatives as antioxidants related to ascorbic acid[J]. J Mol Model, 2020, 26(11):318.
doi: 10.1007/s00894-020-04580-5 URL |
| [6] | Bochot C, Gouron A, Bubacco L, et al. Probing kojic acid binding to tyrosinase enzyme:insights from a model complex and QM/MM calculations[J]. Chem Commun(Camb), 2014, 50(3):308-310. |
| [7] | 蒋利亚. 米曲霉产曲酸菌种选育、发酵条件及抑菌研究[D]. 湖南农业大学, 2012. |
| Jiang LY. Breeding, fermentation conditions and bacteriostasis of Aspergillus oryzae kojic acid producing strain[D]. Changsha: Hunan Agricultural University, 2012. | |
| [8] | Lima GDS, Andrade GF, da SMAN, et al. Novel kojic acid-based functionalized silica nanoparticles for tyrosinase and ache inhibition and antimicrobial applications[J]. Chem Eng Trans CET J, 2018, 64:175-180. |
| [9] | 陆正清, 王艳. 曲酸的发酵法生产及其在食品加工中的应用[J]. 中国调味品, 2008, 33(1):65-67, 71. |
| Lu ZQ, Wang Y. Production of kojic acid by fermentation and its application in food processing[J]. China Condiment, 2008, 33(1):65-67, 71. | |
| [10] |
Yang G, Liu GL, Wang SJ, et al. Pullulan biosynthesis in yeast-like fungal cells is regulated by the transcriptional activator Msn2 and cAMP-PKA signaling pathway[J]. Int J Biol Macromol, 2020, 157:591-603.
doi: 10.1016/j.ijbiomac.2020.04.174 URL |
| [11] |
Zhang M, Gao ZC, Chi Z, et al. cAMP-PKA and HOG1 signaling pathways regulate liamocin production by different ways via the transcriptional activator Msn2 in Aureobasidium melanogenum[J]. Enzyme Microb Technol, 2021, 143:109705.
doi: 10.1016/j.enzmictec.2020.109705 URL |
| [12] | Vamvakas SS, Kapolos J, Farmakis L, et al. Ser625 of msn2 transcription factor is indispensable for ethanol tolerance and alcoholic fermentation process[J]. Biotechnol Prog, 2019, 35(5):e2837. |
| [13] |
Boy-Marcotte E, Perrot M, Bussereau F, et al. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae[J]. J Bacteriol, 1998, 180(5):1044-1052.
pmid: 9495741 |
| [14] |
Chen B, Tesker M, Melamed-Kadosh D, et al. Hog1-induced transcription of RTC3 and HSP12 is robust and occurs in cells lacking Msn2, Msn4, Hot1 and Sko1[J]. PLoS One, 2020, 15(8):e0237540.
doi: 10.1371/journal.pone.0237540 URL |
| [15] |
Chang PK, Scharfenstein LL, Luo M, et al. Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid[J]. Toxins, 2011, 3(1):82-104.
doi: 10.3390/toxins3010082 URL |
| [16] | 吕润玲. 玉米大斑病菌StHog1对转录因子StMsn2的调控作用及StMsn2下游靶基因的筛选与验证[D]. 保定: 河北农业大学, 2019. |
| Lv RL. The regulation of StHog1 on the transcription factor StMsn2 and the screening and verification of the downstream target genes of StMsn2[D]. Baoding: Hebei Agricultural University, 2019. | |
| [17] | 毕付提, 史亚楠, 张久祎, 等. 米曲霉3. 042尿苷/尿嘧啶营养缺陷型遗传转化体系的构建[J]. 食品研究与开发, 2021, 42(3):189-195. |
| Bi FT, Shi YN, Zhang JY, et al. Construction of Aspergillus oryzae 3. 042 uridine/uracil auxotrophy genetic transformation system[J]. Food Res Dev, 2021, 42(3):189-195. | |
| [18] | 曹张磊, 王德培, 张岚. 提高根癌农杆菌介导黑曲霉转化效率的研究[J]. 天津科技大学学报, 2016, 31(2):20-25. |
| Cao ZL, Wang DP, Zhang L. Improvement of transformation efficiency of Aspergillus niger mediated by Agrobacterium tumefaciens[J]. J Tianjin Univ Sci Technol, 2016, 31(2):20-25. | |
| [19] | 鲍龙飞, 秦玉琪, 曲音波. FluG-BrlA途径参与构巢曲霉无性发育机制的研究进展[J]. 微生物学通报, 2014, 41(1):104-110. |
| Bao LF, Qin YQ, Qu YB. Advances in studies of FluG-BrlA pathway involved in asexual developmental mechanisms of Aspergillus nidulans[J]. Microbiol China, 2014, 41(1):104-110. | |
| [20] | 王丽, 向小洪. 海藻糖生物合成途径作为新型抗真菌药物靶标的机遇与挑战[J]. 国外医药:抗生素分册, 2018, 39(5):397-403. |
| Wang L, Xiang XH. Opportunities and challenges of trehalose biosynthesis pathway as a new antifungal drug target[J]. World Notes Antibiot, 2018, 39(5):397-403. | |
| [21] |
Cairns TC, Feurstein C, Zheng XM, et al. Functional exploration of co-expression networks identifies a nexus for modulating protein and citric acid titres in Aspergillus niger submerged culture[J]. Fungal Biol Biotechnol, 2019, 6:18.
doi: 10.1186/s40694-019-0081-x URL |
| [22] |
Liu Q, Ying SH, Li JG, et al. Insight into the transcriptional regulation of Msn2 required for conidiation, multi-stress responses and virulence of two entomopathogenic fungi[J]. Fungal Genet Biol, 2013, 54:42-51.
doi: 10.1016/j.fgb.2013.02.008 URL |
| [23] | 陈浩宇. 黑曲霉中veA及laeA基因功能的研究[D]. 天津: 天津科技大学, 2018. |
| Chen HY. The functional analysis of veA and laeA in Aspergillus niger[D]. Tianjin: Tianjin University of Science & Technology, 2018. | |
| [24] |
Oda K, Kobayashi A, Ohashi S, et al. Aspergillus oryzae laeA regulates kojic acid synthesis genes[J]. Biosci Biotechnol Biochem, 2011, 75(9):1832-1834.
doi: 10.1271/bbb.110235 URL |
| [25] |
Bok JW, Hoffmeister D, Maggio-Hall LA, et al. Genomic mining for Aspergillus natural products[J]. Chem Biol, 2006, 13(1):31-37.
doi: 10.1016/j.chembiol.2005.10.008 URL |
| [26] |
Marui J, Yamane N, Ohashi-Kunihiro S, et al. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)2Cys6transcriptional activator and induced by kojic acid at the transcriptional level[J]. J Biosci Bioeng, 2011, 112(1):40-43.
doi: 10.1016/j.jbiosc.2011.03.010 URL |
| [1] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| [2] | 燕炯, 冯晨毅, 高学坤, 许祥, 杨佳敏, 陈朝阳. 基于CRISPR/Cas9技术构建Plin1基因敲除小鼠模型及表型分析[J]. 生物技术通报, 2022, 38(3): 173-180. |
| [3] | 钟菁, 孙玲玲, 张姝, 蒙园, 支怡飞, 涂黎晴, 徐天鹏, 濮黎萍, 陆阳清. 应用CRISPR/Cas9技术敲除Mda5基因对新城疫及传染性法氏囊病毒复制的影响[J]. 生物技术通报, 2022, 38(11): 90-96. |
| [4] | 王海杰, 王成稷, 郭洋, 王云, 陈艳娟, 梁敏, 王珏, 龚慧, 沈如凌. 基于CRSIPR/Cas9技术构建凝血因子8基因敲除小鼠模型及表型验证[J]. 生物技术通报, 2022, 38(10): 273-280. |
| [5] | 孙靖雅, 马玉超. 假单胞菌Tw224抗砷基因簇的功能研究[J]. 生物技术通报, 2022, 38(1): 141-149. |
| [6] | 王睿, 韩烈保. CRISPR/Cas9介导的二穗短柄草bdfls2敲除突变体的获得[J]. 生物技术通报, 2022, 38(1): 70-76. |
| [7] | 蒋成辉, 曾巧英, 王萌, 潘阳阳, 刘旭明, 尚天甜. CRISPR/Cas9构建srtA基因敲除的金黄色葡萄球菌[J]. 生物技术通报, 2020, 36(9): 253-265. |
| [8] | 梅芬, 李瑞玮, 张娟娟, 左荣霞, 邹云莲, 沈涛, 撒亚莲. 构建基于CRISPR/Cas9技术敲除ALOX5的质粒[J]. 生物技术通报, 2020, 36(3): 110-114. |
| [9] | 王松, 李鹏程, 白朝辉, 许宏鑫, 应研敏, 张墨, 白义春. 利用CRISPR/Cas9技术构建大鼠L2细胞α-ENaC基因敲除稳定细胞株[J]. 生物技术通报, 2020, 36(3): 115-123. |
| [10] | 叶洲杰, 王心睿. CRISPR系统转化医学研究进展[J]. 生物技术通报, 2020, 36(11): 188-197. |
| [11] | 吴航, 李智强, 刘文德. 稻瘟病菌分泌蛋白MoCDIE2诱导植物细胞死亡的功能分析[J]. 生物技术通报, 2020, 36(1): 15-22. |
| [12] | 陈和锋, 朱晁谊, 李爽. 产瓦伦西亚烯酿酒酵母的表达载体适配及发酵碳氮源优化[J]. 生物技术通报, 2020, 36(1): 209-219. |
| [13] | 杨雷, 叶洲杰, 李兆龙, 沈阳坤, 傅雅娟. 利用电转的方法对T细胞TET2基因敲除并探讨TET2对T细胞增殖的影响[J]. 生物技术通报, 2020, 36(1): 229-237. |
| [14] | 吴琴琴, 孙敏, 陈雨, 付雅琴, 曾斌, 贺斌. 米曲霉功能基因组研究策略和进展[J]. 生物技术通报, 2019, 35(8): 186-192. |
| [15] | 赵雅童, 何光明, 瓮茹茹, 石爱琴, 路福平, 李玉. 甘露醇磷酸化酶基因的敲除对D-甘露醇合成的影响[J]. 生物技术通报, 2019, 35(5): 118-124. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||