








生物技术通报 ›› 2022, Vol. 38 ›› Issue (1): 70-76.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0326
收稿日期:2021-03-16
出版日期:2022-01-26
发布日期:2022-02-22
作者简介:王睿,男,博士研究生,研究方向:草地植物分子生物学;E-mail: 基金资助:Received:2021-03-16
Published:2022-01-26
Online:2022-02-22
摘要:
FLS2是一类在植物中保守存在的可识别细菌鞭毛蛋白并激活位于植物先天免疫反应第一层面的重要的植物模式识别受体(pattern recognition receptors,PRRs)。为了进一步研究草坪草植物的先天免疫,本研究以冷季型草坪草模式植物二穗短柄草(Brachypodium distachyon)为材料,利用CRISPR/Cas9基因编辑技术对植物抗病免疫相关的重要基因BdFLS2进行了定向的基因编辑,获得了bdfls2敲除突变体,为草坪草植物的FLS2相关的先天免疫的进一步研究奠定了材料基础。筛选转基因阳性植株,测序分析bdfls2 突变基因,结果显示该突变体中bdfls2 基因编码序列由于碱基的缺失导致提前终止。同时该突变体在对应病原相关分子模式(PAMPs:pathogen-associated molecular pattern)flg22处理后相比于野生型,无显著的ROS爆发或防卫基因的激活,暗示二穗短柄草FLS2能够识别病原相关分子模式flg22激活分子模式触发的免疫(pattern-triggered immunity,PTI)。
王睿, 韩烈保. CRISPR/Cas9介导的二穗短柄草bdfls2敲除突变体的获得[J]. 生物技术通报, 2022, 38(1): 70-76.
WANG Rui, HAN Lie-bao. Generation of bdfls2-knockout Mutant in Brachypodium distachyon Mediated by CRISPR/Cas9[J]. Biotechnology Bulletin, 2022, 38(1): 70-76.
| 组分 Component | 体积 Volume/μL | 反应条件 Reaction conditions |
|---|---|---|
| 退火后的靶点引物 | 2 | 5 h at 37℃ 5 min at 50℃ 10 min at 80℃ |
| pHUE411 载体(约100 ng/μL) | 2 | |
| 10× T4 DNA Ligase Buffer(NEB) | 1.5 | |
| 10× BSA | 1.5 | |
| Bsa I(NEB) | 1 | |
| T4 DNA Ligase(NEB) | 1 | |
| ddH2O | 6 |
表1 酶切-连接体系
Table 1 Enzyme digestion-connection system
| 组分 Component | 体积 Volume/μL | 反应条件 Reaction conditions |
|---|---|---|
| 退火后的靶点引物 | 2 | 5 h at 37℃ 5 min at 50℃ 10 min at 80℃ |
| pHUE411 载体(约100 ng/μL) | 2 | |
| 10× T4 DNA Ligase Buffer(NEB) | 1.5 | |
| 10× BSA | 1.5 | |
| Bsa I(NEB) | 1 | |
| T4 DNA Ligase(NEB) | 1 | |
| ddH2O | 6 |
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| BdPR2 -F | AGCCATCCAGCTCAACTAC |
| BdPR2 -R | CCTTGCCAACATGGTCAATC |
| BdChit8 -F | CTGCTTCAAGGAGGAGATAAAC |
| BdChit8 -R | TCATCCAGAACCACATGGC |
| BdAct -F | GCTGGGCGTGACCTAACTGAC |
| BdAct -R | ATGAAAGATGGCTGGAAAAGGACT |
表2 qRT-PCR引物序列
Table 2 Sequence of primer for qRT-PCR
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| BdPR2 -F | AGCCATCCAGCTCAACTAC |
| BdPR2 -R | CCTTGCCAACATGGTCAATC |
| BdChit8 -F | CTGCTTCAAGGAGGAGATAAAC |
| BdChit8 -R | TCATCCAGAACCACATGGC |
| BdAct -F | GCTGGGCGTGACCTAACTGAC |
| BdAct -R | ATGAAAGATGGCTGGAAAAGGACT |
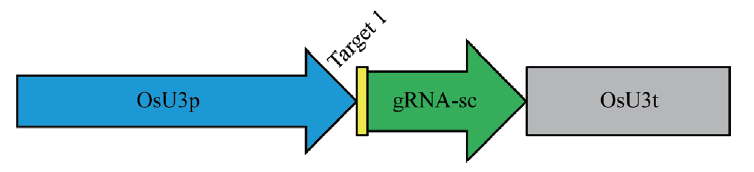
图1 二穗短柄草CRISPR/Cas9-bdfls2基因编辑载体示意图 OsU3:sgRNA-Cas9-pHUE411 载体示意图。浅蓝色部分OsU3p表示水稻U3启动子;黄色和绿色部分表示sgRNA序列;其中黄色部分表示基因的靶序列;灰色部分表示水稻OsU3t终止子
Fig. 1 Illustration of CRISPR/Cas9-bdfls2 gene editing vector in B. distachyon OsU3:sgRNA-Cas9-pHUE411 vector. Light blue indicates the Oryzae U3 promoter. Yellow and green portions indicate the sgRNA sequence. The yellow portion indicates the target sequence of the gene. The grey portion indicates the Oryzae U3 terminator
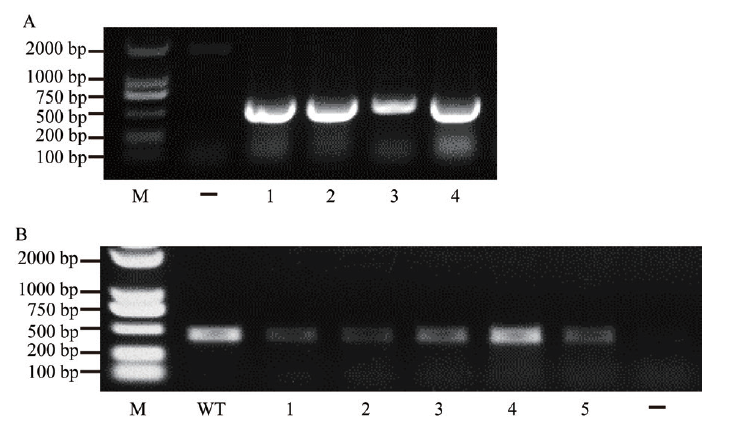
图2 编辑载体和编辑后靶基因的鉴定 A:CRISPR/Cas9-bdfls2基因编辑载体的鉴定,1-4为检测的已转化CRISPR/Cas9-bdfls2的农杆菌菌株;B:靶基因扩增鉴定,1-5为由转基因植株提取的基因组DNA扩增的含有靶基因编辑位点的片段。M为DNA marker;WT为二穗短柄草Bd21野生型对照;-为阴性对照
Fig. 2 Identification of gene editing vector and edited target gene A:Identification of CRISPR/Cas9-bdfls2 gene editing vector. 1-4 are the assayed Agrobacterium tumefaciens strains for transformed CRISPR/Cas9-bdfls2. B:Amplification and identification of target gene. 1-5 are genome-DNA-amplified fragments containing target gene editing sites extracted from transgenic plants. M is the DNA marker. Bd21 is the wild type(WT). - indicates the negative control
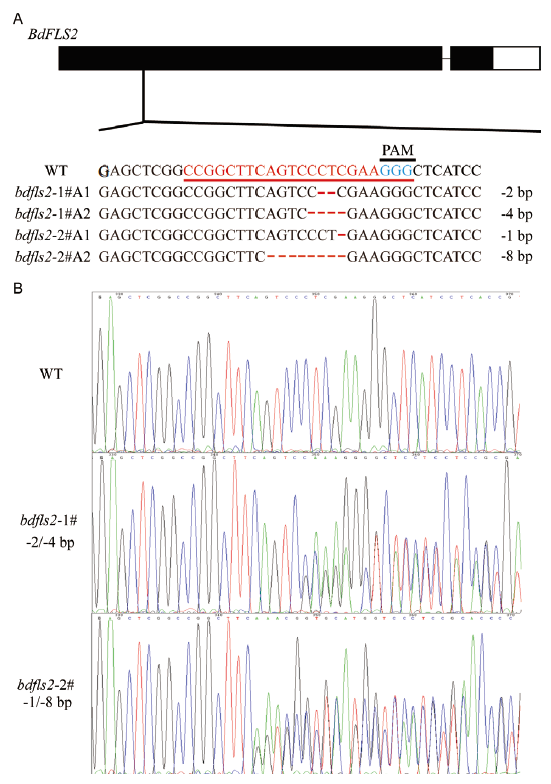
图3 二穗短柄草bdfls2 突变体基因型检测 A:bdfls2突变体的基因型。红色下划线表示sgRNA:Cas9编辑的靶序列,蓝色代表对应的前间隔序列邻近基序(PAM),1#、2#分别代表不同的突变体植株,A1、A2分别代表Allele 1和Allele 2,序列中红色 - 表示碱基缺失;B:bdfls2突变体的的测序图。WT为作为对照的Bd21野生型
Fig. 3 Genotyping of bdfls2 mutants in B. distachyon A:Genotyping of bdfls2 mutants. Red underline refers to sgRNA:Cas9-edited bdfls2 mutation. Blue underline refer to schematic illustration of the sgRNA:Cas9 targets and the corresponding protospacer-adjacent motif(PAM)(blue)of the targeted genes. The 1# and 2# stands for different mutant plants,A1 and A2 for Allele 1 and Allele 2,and the red lines in sequences stands for base deletion. B:Sequencing of bdfls2 mutants. WT is the Bd21 wild type as control
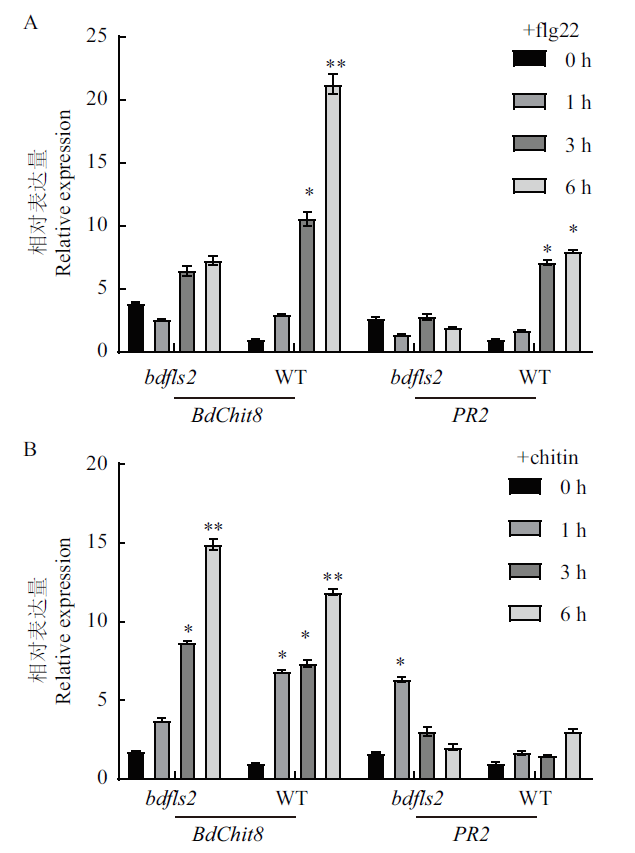
图5 实时荧光定量RT-PCR 对PAMPs诱导的防卫基因的检测 二穗短柄草叶片在5 μmol/L flg22(A)或 100 μg/mL chitin(B)未处理(0 h)及处理后1,3,6 h的 BdChit8和PR2 基因的相对表达量,通过实时荧光定量RT-PCR检测得出。图中各组数据均有3组生物学重复,各柱表示为平均值±标准误。图中柱子上的*或**分别代表P < 0.05 或P < 0.01
Fig. 5 Detection of defense gene expression induced upon different PAMPs treatment by real-time quantitative RT-PCR The relative BdChit8 and PR2 expression in the leaves of B. distachyon seedlings treated with 5 μmol/L flg22(A)or 100 μg/mL chitin(B)were determined by quantitative real-time PCR. Data are presented as the mean ± standard error of three biological replicates.* or ** above the bars indicate P < 0.05 or P < 0.01
| [1] |
Ausubel FM. Are innate immune signaling pathways in plants and animals conserved?[J]. Nat Immunol, 2005, 6(10):973-979.
doi: 10.1038/ni1253 URL |
| [2] |
Jones JDG, Dangl JL. The plant immune system[J]. Nature, 2006, 444(7117):323-329.
doi: 10.1038/nature05286 URL |
| [3] |
Sanabria N, Goring D, Nürnberger T, et al. Self/nonself perception and recognition mechanisms in plants:a comparison of self-incompatibility and innate immunity[J]. New Phytol, 2008, 178(3):503-514.
doi: 10.1111/j.1469-8137.2008.02403.x pmid: 18346103 |
| [4] |
Zipfel C. Plant pattern-recognition receptors[J]. Trends Immunol, 2014, 35(7):345-351.
doi: 10.1016/j.it.2014.05.004 pmid: 24946686 |
| [5] |
Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity(PTI)[J]. Mol Plant, 2015, 8(4):521-539.
doi: 10.1016/j.molp.2014.12.022 pmid: 25744358 |
| [6] |
Zipfel C. Pattern-recognition receptors in plant innate immunity[J]. Curr Opin Immunol, 2008, 20(1):10-16.
doi: 10.1016/j.coi.2007.11.003 pmid: 18206360 |
| [7] |
Gómez-Gómez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana[J]. Plant J, 1999, 18(3):277-284.
pmid: 10377993 |
| [8] |
Robatzek S, Bittel P, Chinchilla D, et al. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities[J]. Plant Mol Biol, 2007, 64(5):539-547.
pmid: 17530419 |
| [9] |
Huang PY, Yeh YH, Liu AC, et al. The Arabidopsis LecRK-VI. 2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity[J]. Plant J, 2014, 79(2):243-255.
doi: 10.1111/tpj.12557 URL |
| [10] |
Gómez-Gómez L, Boller T. FLS2:an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis[J]. Mol Cell, 2000, 5(6):1003-1011.
pmid: 10911994 |
| [11] |
Takai R, Isogai A, Takayama S, et al. Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice[J]. Mol Plant Microbe Interact, 2008, 21(12):1635-1642.
doi: 10.1094/MPMI-21-12-1635 URL |
| [12] |
Hann DR, Rathjen JP. Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana[J]. Plant J, 2007, 49(4):607-618.
doi: 10.1111/j.1365-313X.2006.02981.x URL |
| [13] |
Boller T, Felix G. A renaissance of elicitors:perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors[J]. Annu Rev Plant Biol, 2009, 60:379-406.
doi: 10.1146/arplant.2009.60.issue-1 URL |
| [14] |
Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and Archaea[J]. Nature, 2012, 482(7385):331-338.
doi: 10.1038/nature10886 URL |
| [15] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6):1262-1278.
doi: 10.1016/j.cell.2014.05.010 URL |
| [16] |
Yin K, Gao C, Qiu JL. Progress and prospects in plant genome editing[J]. Nat Plants, 2017, 3:17107.
doi: 10.1038/nplants.2017.107 URL |
| [17] |
Ji X, Wang D, Gao C. CRISPR editing-mediated antiviral immunity:a versatile source of resistance to combat plant virus infections[J]. Sci China Life Sci, 2019, 62(9):1246-1249.
doi: 10.1007/s11427-019-9722-2 URL |
| [18] |
Kellogg EA. Evolutionary history of the grasses[J]. Plant Physiol, 2001, 125(3):1198-1205.
pmid: 11244101 |
| [19] |
Davis JI, Soreng RJ. Phylogenetic structure in the grass family(Poaceae)as inferred from chloroplast DNA restriction site variation[J]. Am J Bot, 1993, 80(12):1444.
doi: 10.1002/ajb2.1993.80.issue-12 URL |
| [20] |
Liu H, Ding Y, Zhou Y, et al. CRISPR-P 2. 0:an improved CRISPR-Cas9 tool for genome editing in plants[J]. Mol Plant, 2017, 10(3):530-532.
doi: 10.1016/j.molp.2017.01.003 URL |
| [21] |
Chinchilla D, Bauer Z, Regenass M, et al. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception[J]. Plant Cell, 2006, 18(2):465-476.
pmid: 16377758 |
| [22] |
Xing HL, Dong L, Wang ZP, et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants[J]. BMC Plant Biol, 2014, 14:327.
doi: 10.1186/s12870-014-0327-y URL |
| [23] | 吴雪莉, 刘金星, Nielsen K, 等. 二穗短柄草幼胚再生体系及农杆菌介导转化的初步研究[J]. 草业学报, 2010, 19(5):9-16. |
| Wu XL, Liu JX, Nielsen K, et al. Agrobacterium-mediated transformation of Brachypodium distachyon through embryogenic calli derived from immature embryos[J]. Acta Prataculturae Sin, 2010, 19(5):9-16. | |
| [24] |
Guo W, Zuo ZL, Cheng X, et al. The chloride channel family gene CLCd negatively regulates pathogen-associated molecular pattern(PAMP)-triggered immunity in Arabidopsis[J]. J Exp Bot, 2014, 65(4):1205-1215.
doi: 10.1093/jxb/ert484 URL |
| [25] |
Pérez FJ, Rubio S. An improved chemiluminescence method for hydrogen peroxide determination in plant tissues[J]. Plant Growth Regul, 2006, 48(1):89-95.
doi: 10.1007/s10725-005-5089-y URL |
| [26] |
Liu W, Xie X, Ma X, et al. DSDecode:a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations[J]. Mol Plant, 2015, 8(9):1431-1433.
doi: 10.1016/j.molp.2015.05.009 URL |
| [27] |
Wang S, Sun Z, Wang H, et al. Rice OsFLS2-mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola[J]. Mol Plant, 2015, 8(7):1024-1037.
doi: 10.1016/j.molp.2015.01.012 URL |
| [28] |
Fürst U, Zeng Y, Albert M, et al. Perception of Agrobacterium tumefaciens flagellin by FLS2XL confers resistance to crown gall disease[J]. Nat Plants, 2020, 6(1):22-27.
doi: 10.1038/s41477-019-0578-6 URL |
| [1] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [2] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [3] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [4] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [5] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [6] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [7] | 王兵, 赵会纳, 余婧, 陈杰, 骆梅, 雷波. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控[J]. 生物技术通报, 2023, 39(10): 197-208. |
| [8] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [9] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [10] | 史亚楠, 王德培, 王一川, 周昊, 薛鲜丽. 敲除msn2对米曲霉生长和发酵产曲酸的影响[J]. 生物技术通报, 2022, 38(8): 188-197. |
| [11] | 刘静静, 刘晓蕊, 李琳, 王盈, 杨海元, 戴一凡. 利用CRISPR/Cas9技术建立OXTR基因敲除猪胎儿成纤维细胞系[J]. 生物技术通报, 2022, 38(6): 272-278. |
| [12] | Olalekan Amoo, 胡利民, 翟云孤, 范楚川, 周永明. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105. |
| [13] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| [14] | 燕炯, 冯晨毅, 高学坤, 许祥, 杨佳敏, 陈朝阳. 基于CRISPR/Cas9技术构建Plin1基因敲除小鼠模型及表型分析[J]. 生物技术通报, 2022, 38(3): 173-180. |
| [15] | 钟菁, 孙玲玲, 张姝, 蒙园, 支怡飞, 涂黎晴, 徐天鹏, 濮黎萍, 陆阳清. 应用CRISPR/Cas9技术敲除Mda5基因对新城疫及传染性法氏囊病毒复制的影响[J]. 生物技术通报, 2022, 38(11): 90-96. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
