








生物技术通报 ›› 2022, Vol. 38 ›› Issue (11): 49-57.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1602
收稿日期:2021-12-30
出版日期:2022-11-26
发布日期:2022-12-01
作者简介:孙忠娟,女,硕士研究生,研究方向:真菌学;E-mail:基金资助:
SUN Zhong-juan( ), LIU Qian-qian, GUO Yu-qian, WANG Guang-hui(
), LIU Qian-qian, GUO Yu-qian, WANG Guang-hui( ), WANG Chen-fang(
), WANG Chen-fang( )
)
Received:2021-12-30
Published:2022-11-26
Online:2022-12-01
摘要:
常用的基因敲除技术无法应用于某些特殊激酶的研究,旨为在植物病原真菌中建立基于蛋白激酶人工改造的analog-sensitive蛋白激酶研究系统,为蛋白激酶的研究提供新的思路和方法。采用网站预测蛋白激酶的“守门员”残基,用载体回补法对其进行突变获得相应的类似物敏感型(analog-sensitive,as)突变体gpmk1-as与pmk1-as,并检测类似物敏感型蛋白激酶的功能及其对ATP类似物抑制剂1-NM-PP1的敏感性。结果显示,Gpmk1与Pmk1的守门员残基分别为Q103和Q104,此位点在多个代表性真菌中均十分保守;类似物敏感型突变体gpmk1-as的菌落生长速度与野生型PH-1一致,均快于Gpmk1敲除突变体,Pmk1-as能够产生与野生型Guy11一样成熟的黑化附着孢,而Pmk1敲除突变体无法产生;在5 μmol/L的1-NM-PP1条件下,gpmk1-as的菌落生长速度下降,但PH-1正常生长,在10 μmol/L的1-NM-PP条件下,pmk1-as无法形成附着孢,但Guy11可以形成成熟的黑化附着孢。结果表明,Gpmk1和Pmk1的类似物敏感型蛋白激酶均能行使正常的蛋白功能,却对ATP类似物抑制剂1-NM-PP1十分敏感。
孙忠娟, 刘倩倩, 郭雨纤, 王光辉, 王晨芳. Analog-sensitive蛋白激酶研究系统在植物病原真菌中的建立[J]. 生物技术通报, 2022, 38(11): 49-57.
SUN Zhong-juan, LIU Qian-qian, GUO Yu-qian, WANG Guang-hui, WANG Chen-fang. Establishment of Analog-sensitive Protein Kinase Research System in Plant Pathogenic Fungi[J]. Biotechnology Bulletin, 2022, 38(11): 49-57.
| 引物名称Primer | 引物序列Sequence(5'-3') |
|---|---|
| GPMK1-as/1F | AGGGAACAAAAGCTGGGTACCGGGGTATGGT- GGTCAAGGTTATG |
| GPMK1-as/2R | ACAGTGGCGTTTCGTACTCCGATCAGATAGAC |
| GPMK1-as/3F | GTCTATCTGATCGGAGTACGAAACGCCACTGT |
| GPMK1-as/4R | GATTTCAGTAACGTTAAGTGGATCCATGGTGG- CAAGTAAAGGAATGAG |
| GPMK1-as/5F | CTGTATCCGTAGATGGGAGGTA |
| GPMK1-as/6F | GACCGTGCTTGCTGTATCCT |
| GPMK1-as/7F | GACCGAACGACCGATTGACT |
| GPMK1-as/8F | CAACCTCCTCCTCAACGCCAACT |
| GPMK1-as/9F | ACCCATACCTTGAGCCTTACC |
| PMK1-as/1F | AGGGAACAAAAGCTGGGTACCGCGTCAAGCC- ATCGCGTTATCT |
| PMK1-as/2R | GAGTGTTCAGCAGCAACGGACCCCGATCAGG- TACACCTCGTT |
| PMK1-as/3F | TTCAACGAGGTGTACCTGATCGGGGTCCGTTG- CTGCTGAACACTCC |
| PMK1-as/4R | GATTTCAGTAACGTTAAGTGGATCCGGCACAG- CCAATCCACGAATACACA |
| PMK1-as/5F | ATTCTCGTTGGTCCATCCGTC |
| PMK1-as/6F | TCCACCAAGCAACTCTTCGGG |
| PMK1-as/7F | TCAAGGCAATGCACTCGGCAAAC |
| PMK1-as/8F | GTACCATGACCCCGATGATGAACC |
| PMK1-as/9F | CGGCACTCACAATTAAGCCGTTG |
| GPMK1-5F | CAACTGTGATCTCAAGGTCTGCG |
| GPMK1-6R | TGCTTCAGAGCTTCCTCCACAG |
| PMK1-5F | CGAAACGCCGTCTACTGTACTTG |
| PMK1-6R | CAGTACTGGGGAGAACATCGTG |
表1 本实验所用的引物序列
Table 1 Primer sequences used in this study
| 引物名称Primer | 引物序列Sequence(5'-3') |
|---|---|
| GPMK1-as/1F | AGGGAACAAAAGCTGGGTACCGGGGTATGGT- GGTCAAGGTTATG |
| GPMK1-as/2R | ACAGTGGCGTTTCGTACTCCGATCAGATAGAC |
| GPMK1-as/3F | GTCTATCTGATCGGAGTACGAAACGCCACTGT |
| GPMK1-as/4R | GATTTCAGTAACGTTAAGTGGATCCATGGTGG- CAAGTAAAGGAATGAG |
| GPMK1-as/5F | CTGTATCCGTAGATGGGAGGTA |
| GPMK1-as/6F | GACCGTGCTTGCTGTATCCT |
| GPMK1-as/7F | GACCGAACGACCGATTGACT |
| GPMK1-as/8F | CAACCTCCTCCTCAACGCCAACT |
| GPMK1-as/9F | ACCCATACCTTGAGCCTTACC |
| PMK1-as/1F | AGGGAACAAAAGCTGGGTACCGCGTCAAGCC- ATCGCGTTATCT |
| PMK1-as/2R | GAGTGTTCAGCAGCAACGGACCCCGATCAGG- TACACCTCGTT |
| PMK1-as/3F | TTCAACGAGGTGTACCTGATCGGGGTCCGTTG- CTGCTGAACACTCC |
| PMK1-as/4R | GATTTCAGTAACGTTAAGTGGATCCGGCACAG- CCAATCCACGAATACACA |
| PMK1-as/5F | ATTCTCGTTGGTCCATCCGTC |
| PMK1-as/6F | TCCACCAAGCAACTCTTCGGG |
| PMK1-as/7F | TCAAGGCAATGCACTCGGCAAAC |
| PMK1-as/8F | GTACCATGACCCCGATGATGAACC |
| PMK1-as/9F | CGGCACTCACAATTAAGCCGTTG |
| GPMK1-5F | CAACTGTGATCTCAAGGTCTGCG |
| GPMK1-6R | TGCTTCAGAGCTTCCTCCACAG |
| PMK1-5F | CGAAACGCCGTCTACTGTACTTG |
| PMK1-6R | CAGTACTGGGGAGAACATCGTG |
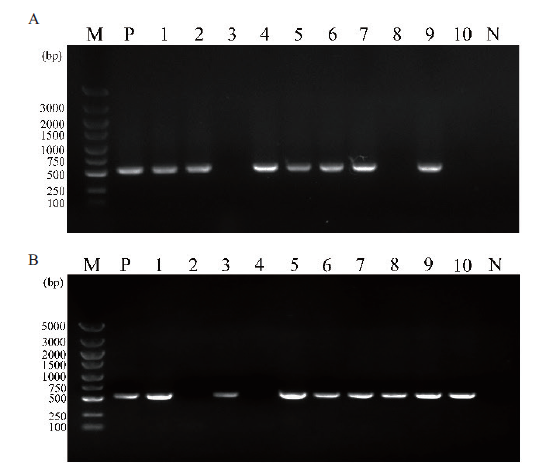
图4 gpmk1-as(A)及pmk1-as(B)转化子的PCR检测 M:DNA marker;P:阳性对照;N:阴性对照
Fig.4 PCR detection of gpmk1-as(A)and pmk1-as(B)transformants M:DNA marker;P:positive control;N:negative control
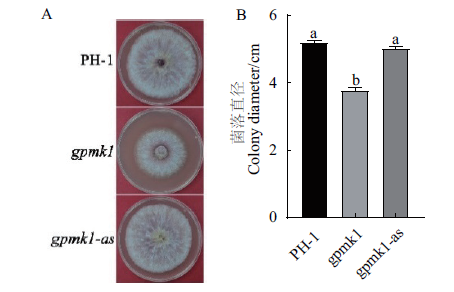
图5 菌株PH-1、gpmk1和gpmk1-as在PDA平板上生长2 d的菌落(A)及菌落直径统计分析(B) 经单因素方差分析及Duncan多极差检验,不同字母表示差异显著(P=0.05)
Fig.5 Cultures of the strains PH-1,gpmk1 and gpmk1-as grown on PDA plate for 2 d(A)and colony dia-meters of the indicated strains(B) Different letters indicate significant differences based on ANOVA analysis followed by Duncan’s multiple range test(P=0.05)
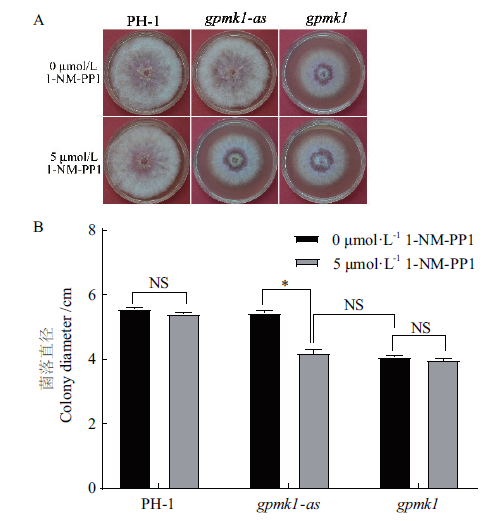
图7 1-NM-PP1处理后菌株PH-1、gpmk1-as和gpmk1在PDA平板上生长2 d的菌落(A)及直径统计分析(B) 经t检验分析(P=0.05)
Fig.7 Cultures of the strain PH-1,gpmk1 and gpmk1-as grown on PDA plate for 2 d after 1-NM-PP1 treatm-ent(A)and colony diameters of the indicated str-ains(B) Significant differences based on t test(P=0.05)
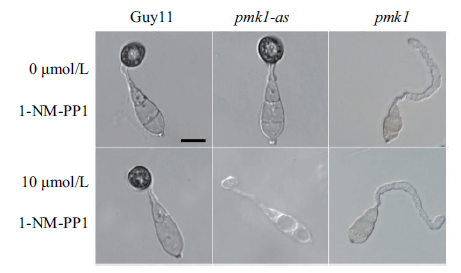
图8 1-NM-PP1处理后菌株Guy11、pmk1和pmk1-as在疏水玻片上的附着孢形成(Bar=50 μm)
Fig.8 Appressorium formation of strain Guy11,pmk1 and pmk1-as on hydrophobic slides after 1-NM-PP1 treatment(Bar=50 μm)
| [1] | 宋嘉宁, MUHAMMAD Tahir, 张文婷, 等. 自噬相关蛋白激酶的研究进展[J]. 河北师范大学学报:自然科学版, 2021, 45(6):612-619. |
| Song JN, Tahir M, Zhang WT, et al. Research progress of autophagy-associated protein kinases[J]. J Hebei Norm Univ Nat Sci, 2021, 45(6):612-619. | |
| [2] |
Turrà D, Segorbe D, di Pietro A. Protein kinases in plant-pathogenic fungi:conserved regulators of infection[J]. Annu Rev Phytopathol, 2014, 52:267-288.
doi: 10.1146/annurev-phyto-102313-050143 URL |
| [3] |
Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update[J]. Trends Biochem Sci, 2000, 25(12):596-601.
pmid: 11116185 |
| [4] | Jiang C, Zhang X, Liu HQ, et al. Mitogen-activated protein kinase signaling in plant pathogenic fungi[J]. PLoS Pathog, 2018, 14(3):e1006875. |
| [5] | Wang CF, Zhang SJ, Hou R, et al. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum[J]. PLoS Pathog, 2011, 7(12):e1002460. |
| [6] |
Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe[J]. Mol Gen Genet, 1976, 146(2):167-178.
doi: 10.1007/BF00268085 URL |
| [7] |
Li GT, Zhou XY, Xu JR. Genetic control of infection-related development in Magnaporthe oryzae[J]. Curr Opin Microbiol, 2012, 15(6):678-684.
doi: 10.1016/j.mib.2012.09.004 URL |
| [8] |
Ma ZP, Zhu PP, Shi H, et al. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components[J]. Nature, 2019, 568(7751):259-263.
doi: 10.1038/s41586-019-1057-y URL |
| [9] |
Kawasumi M, Nghiem P. Chemical genetics:elucidating biological systems with small-molecule compounds[J]. J Invest Dermatol, 2007, 127(7):1577-1584.
pmid: 17568801 |
| [10] | Pincus D, Pandey JP, Feder ZA, et al. Engineering allosteric regulation in protein kinases[J]. Sci Signal, 2018, 11(555):eaar3250. |
| [11] |
Gregan J, Zhang C, Rumpf C, et al. Construction of conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe[J]. Nat Protoc, 2007, 2(11):2996-3000.
doi: 10.1038/nprot.2007.447 pmid: 18007635 |
| [12] |
Bishop AC, Ubersax JA, Petsch DT, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase[J]. Nature, 2000, 407(6802):395-401.
doi: 10.1038/35030148 URL |
| [13] |
Olsen LCB, Færgeman NJ. Chemical genomics and emerging DNA technologies in the identification of drug mechanisms and drug targets[J]. Curr Top Med Chem, 2012, 12(12):1331-1345.
pmid: 22690680 |
| [14] |
Bishop AC, Kung CY, Shah K, et al. Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach[J]. J Am Chem Soc, 1999, 121(4):627-631.
doi: 10.1021/ja983267v URL |
| [15] |
Verschueren K, Cobbaut M, Demaerel J, et al. Discovery of a potent protein kinase D inhibitor:insights in the binding mode of pyrazolo[3, 4- d]pyrimidine analogues[J]. MedChemComm, 2017, 8(3):640-646.
doi: 10.1039/c6md00675b pmid: 28890776 |
| [16] |
Garske AL, Peters U, Cortesi AT, et al. Chemical genetic strategy for targeting protein kinases based on covalent complementarity[J]. Proc Natl Acad Sci USA, 2011, 108(37):15046-15052.
doi: 10.1073/pnas.1111239108 URL |
| [17] |
Weiss EL, Bishop AC, Shokat KM, et al. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p[J]. Nat Cell Biol, 2000, 2(10):677-685.
doi: 10.1038/35036300 pmid: 11025657 |
| [18] |
Rubenstein EM, McCartney RR, Zhang C, et al. Access denied:Snfdenied:Snf 1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase[J]. J Biol Chem, 2008, 283(1):222-230.
doi: 10.1074/jbc.M707957200 pmid: 17991748 |
| [19] |
Decker TM, Forné I, Straub T, et al. Analog-sensitive cell line identifies cellular substrates of CDK9[J]. Oncotarget, 2019, 10(65):6934-6943.
doi: 10.18632/oncotarget.27334 URL |
| [20] |
Koch A, Hauf S. Strategies for the identification of kinase substrates using analog-sensitive kinases[J]. Eur J Cell Biol, 2010, 89(2/3):184-193.
doi: 10.1016/j.ejcb.2009.11.024 URL |
| [21] |
Harashima H, Dissmeyer N, Hammann P, et al. Modulation of plant growth in vivo and identification of kinase substrates using an analog-sensitive variant of CYCLIN-DEPENDENT KINASE A;1[J]. BMC Plant Biol, 2016, 16(1):209.
pmid: 27669979 |
| [22] |
Zhang X, Wang ZY, Jiang C, et al. Regulation of biotic interactions and responses to abiotic stresses by MAP kinase pathways in plant pathogenic fungi[J]. Stress Biol, 2021, 1(1):1-19.
doi: 10.1007/s44154-021-00001-6 URL |
| [23] | 王光辉. 禾谷镰刀菌AMT1基因的功能研究[D]. 杨凌: 西北农林科技大学, 2010. |
| Wang GH. Functional characterization of AMT1 genes in Fusarium graminearum[D]. Yangling: Northwest A & F University, 2010. | |
| [24] | 王光辉. 稻瘟菌致病相关信号黏蛋白MoMsb2以及转录因子MoWor1功能研究[D]. 杨凌: 西北农林科技大学, 2015. |
| Wang GH. Function analysis of pathogenicity-related signalling mucin MoMsb2 and transcriptional regulator MoWor1 in Magnaporthe oryzae[D]. Yangling: Northwest A & F University, 2015. | |
| [25] |
Bruno KS, Tenjo F, Li L, et al. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea[J]. Eukaryot Cell, 2004, 3(6):1525-1532.
doi: 10.1128/EC.3.6.1525-1532.2004 URL |
| [26] |
Zhao XH, Mehrabi R, Xu JR. Mitogen-activated protein kinase pathways and fungal pathogenesis[J]. Eukaryot Cell, 2007, 6(10):1701-1714.
pmid: 17715363 |
| [27] |
Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron:a method for constructing temperature-sensitive mutants[J]. Science, 1994, 263(5151):1273-1276.
pmid: 8122109 |
| [28] |
Zeidler MP, Tan CG, Bellaiche Y, et al. Temperature-sensitive control of protein activity by conditionally splicing inteins[J]. Nat Biotechnol, 2004, 22(7):871-876.
pmid: 15184905 |
| [29] |
Zhang X, Liu WD, Li Y, et al. Expression of HopAI interferes with MAP kinase signalling in Magnaporthe oryzae[J]. Environ Microbiol, 2017, 19(10):4190-4204.
doi: 10.1111/1462-2920.13884 pmid: 28799700 |
| [30] |
Roskoski R Jr. A historical overview of protein kinases and their targeted small molecule inhibitors[J]. Pharmacol Res, 2015, 100:1-23.
doi: 10.1016/j.phrs.2015.07.010 pmid: 26207888 |
| [31] |
Knight ZA, Shokat KM. Chemical genetics:where genetics and pharmacology meet[J]. Cell, 2007, 128(3):425-430.
doi: 10.1016/j.cell.2007.01.021 URL |
| [32] |
Kim S, Park J, Kim D, et al. Development of a versatile copper-responsive gene expression system in the plant-pathogenic fungus Fusarium graminearum[J]. Mol Plant Pathol, 2021, 22(11):1427-1435.
doi: 10.1111/mpp.13118 URL |
| [1] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [2] | 王伟宸, 赵进, 黄薇颐, 郭芯竹, 李婉颖, 张卓. 芽胞杆菌代谢产物防治三种常见植物病原真菌的研究进展[J]. 生物技术通报, 2023, 39(3): 59-68. |
| [3] | 刘媛媛, 杨冬杰, 左东云, 程海亮, 张友平, 吕丽敏, 王巧连, 宋国立. 棉花GhD6PKL2的克隆及功能验证[J]. 生物技术通报, 2021, 37(8): 111-120. |
| [4] | 王海波, 郭俊云, 田雪莲. 小桐子SnRK1蛋白激酶α亚基基因的克隆及原核表达分析[J]. 生物技术通报, 2019, 35(6): 39-47. |
| [5] | 周利明, 卢鑫蕊, 马声葳, 房玮. 钙依赖蛋白激酶CPK14在花粉管生长中的功能分析[J]. 生物技术通报, 2019, 35(6): 55-61. |
| [6] | 袁敏,齐玉荣,王瑞菊,. 蛋白激酶BIN2的纯化及活性分析[J]. 生物技术通报, 2017, 33(7): 145-149. |
| [7] | 彭卫福, 吴志明, 陈未, 曾勇军, 李昆太. 拮抗多种植物病原真菌Streptomyces triostinicus C2的分离与鉴定[J]. 生物技术通报, 2016, 32(7): 106-111. |
| [8] | 郑超, 李登高, 白薇. 植物富含半胱氨酸的类受体激酶的研究进展[J]. 生物技术通报, 2016, 32(11): 10-17. |
| [9] | 韩坤煌, 戴燕彬, 邹志华, 张子平, 王艺磊. CKS1B参与拟穴青蟹性腺发育的研究[J]. 生物技术通报, 2016, 32(11): 188-193. |
| [10] | 杨小岚, 陈玉婵, 李浩华, 章卫民. 23株海洋真菌的分子鉴定及其抗植物病原真菌和细胞毒活性研究[J]. 生物技术通报, 2014, 0(8): 132-137. |
| [11] | 韩国灿, 姜少杰, 邹飞雁. 酪蛋白激酶Iα与细胞信号通路[J]. 生物技术通报, 2014, 0(3): 22-29. |
| [12] | 夏洪丽,蔡佳, 鲁义善,简纪常, 吴灶和. 红笛鲷p38β MAPK基因的克隆及原核表达[J]. 生物技术通报, 2014, 0(12): 177-183. |
| [13] | 姜珊珊, 张丹, 孔祥培, 周严, 李德全. 植物中的钙依赖蛋白激酶(CDPK)的结构特征和功能研究进展[J]. 生物技术通报, 2013, 0(6): 12-19. |
| [14] | 刘晓妹;蒲金基;张欣;漆艳香;谢艺贤;张贺;郑服丛;. 多主棒孢菌调控致病性相关基因CCK1的克隆及生物信息学分析[J]. , 2012, 0(10): 168-172. |
| [15] | 谢昕;杨英歌;黄继翔;. 植物病原真菌广谱拮抗菌的筛选鉴定及发酵条件初步研究[J]. , 2012, 0(09): 143-148. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||