








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 243-253.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0686
李琦1( ), 杨晓蕾1, 李晓林2, 申友磊1, 李建宏1, 姚拓1(
), 杨晓蕾1, 李晓林2, 申友磊1, 李建宏1, 姚拓1( )
)
收稿日期:2022-06-04
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
姚拓,男,博士,教授,研究方向:草地微生物多样性; E-mail: yaotuo@gsau.edu.cn作者简介:李琦,男,博士研究生,研究方向:溶磷微生物; E-mail: lq15774738520@163.com
基金资助:
LI Qi1( ), YANG Xiao-lei1, LI Xiao-lin2, SHEN You-lei1, LI Jian-hong1, YAO Tuo1(
), YANG Xiao-lei1, LI Xiao-lin2, SHEN You-lei1, LI Jian-hong1, YAO Tuo1( )
)
Received:2022-06-04
Published:2023-03-26
Online:2023-04-10
摘要:
旨为从高寒草地燕麦根际定向筛选解植酸磷微生物资源,筛选促生潜力菌株,分析植酸酶编码基因。采用国际植物研究所磷酸盐生长培养基(NBRIP)分离及筛选菌株,16S rRNA基因鉴定其分类地位,并测定菌株植酸酶活性及促生特性,结合简并PCR和高效热不对称交错PCR(hiTAIL-PCR)扩增植酸酶基因完整序列,并进行生物信息学分析及异源表达。共获得107株菌株,其中51株能在NBRIP培养基上形成清晰溶磷圈,鉴定为2门10科11属,以假单胞菌(Pseudomonas)为优势菌。14株不同种假单胞菌均检测出植酸酶活性,具有溶解有机/无机磷、分泌IAA(3-indoleacetic acid)、固氮及拮抗植物病原菌的促生活性。获得了3株菌株的植酸酶(PHY65、PHY101和PHY131)序列,预测为β-螺旋植酸酶(β-propeller phytases,BPPhy)家族蛋白,其中重组PHY65的比活性为28.2 U/mg。研究结果可为解磷生物菌剂的研发与利用提供优良菌株资源,为植酸酶的生产应用提供理论基础。
李琦, 杨晓蕾, 李晓林, 申友磊, 李建宏, 姚拓. 高寒草地燕麦根际解植酸磷促生菌鉴定及其优势菌假单胞菌属菌株功能特性[J]. 生物技术通报, 2023, 39(3): 243-253.
LI Qi, YANG Xiao-lei, LI Xiao-lin, SHEN You-lei, LI Jian-hong, YAO Tuo. Identification of Phytate Phosphorus-solubilizing PGPB in Avena sativa Rhizosphere from Alpine Grassland and Functional Characteristics of Dominant Genus Pseudomonas sp.[J]. Biotechnology Bulletin, 2023, 39(3): 243-253.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Usage |
|---|---|---|
| B6-5 LB-0a | ATTCACCCTGAGCAGCCCTCTA | GS6-5植酸酶基因两翼序列扩增 |
| B6-5 LB-1a | ACGATGGACTCCAGTCCGGCCCGCGTACTTGGCACCAACAAGAAGCA | |
| B6-5 LB-2a | TCGGGCGCCTCAACAACGTCGATA | |
| B6-5 RB-0a | GACCGCCAGTTCCTGCAACAA | |
| B6-5 RB-1a | ACGATGGACTCCAGTCCGGCCAGGCCCTGCTTCTTGTTGGTGCCA | |
| B6-5 RB-2a | ATCCACACCGCTGGATCATCGGCT | |
| B10-1 LB-0a | GGATTCACCCGCAGCAACCGT | GS10-1植酸酶基因两翼序列扩增 |
| B10-1 LB-1a | ACGATGGACTCCAGTCCGGCCAAGCAGGGACTGATGGCGTACGACC | |
| B10-1 LB-2a | CCGGTGGGGCGCCTGAACAATGTC | |
| B10-1 RB-0a | GCGCAAGTCGACATTGTTCAG | |
| B10-1 RB-1a | ACGATGGACTCCAGTCCGGCCTGCCTTGCAGGTCGTACGCCATCAG | |
| B10-1 RB-2a | GTGCCCAGTACACGGCTCAGCGAC | |
| B13-1 LB-0a | CCGCCGTATGGATTCACCCGCAGCA | GS13-1植酸酶基因两翼序列扩增 |
| B13-1 LB-1a | ACGATGGACTCCAGTCCGGCCCGCTGAGCCGTGTACTGGGCACCAA | |
| B13-1 LB-2a | ACTGCTACAGGAGTTGCCGGTGGGG | |
| B13-1 RB-0a | CGCCCCACCGGCAACTCCTGTAGCA | |
| B13-1 RB-1a | ACGATGGACTCCAGTCCGGCCTTGCAGGTCGTACGCCATCAGTCCC | |
| B13-1 RB-2a | GCGACGGTTGCTGCGGGTGAATCCA |
表1 hiTAIL-PCR引物列表
Table 1 Primers used in hiTAIL-PCR
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Usage |
|---|---|---|
| B6-5 LB-0a | ATTCACCCTGAGCAGCCCTCTA | GS6-5植酸酶基因两翼序列扩增 |
| B6-5 LB-1a | ACGATGGACTCCAGTCCGGCCCGCGTACTTGGCACCAACAAGAAGCA | |
| B6-5 LB-2a | TCGGGCGCCTCAACAACGTCGATA | |
| B6-5 RB-0a | GACCGCCAGTTCCTGCAACAA | |
| B6-5 RB-1a | ACGATGGACTCCAGTCCGGCCAGGCCCTGCTTCTTGTTGGTGCCA | |
| B6-5 RB-2a | ATCCACACCGCTGGATCATCGGCT | |
| B10-1 LB-0a | GGATTCACCCGCAGCAACCGT | GS10-1植酸酶基因两翼序列扩增 |
| B10-1 LB-1a | ACGATGGACTCCAGTCCGGCCAAGCAGGGACTGATGGCGTACGACC | |
| B10-1 LB-2a | CCGGTGGGGCGCCTGAACAATGTC | |
| B10-1 RB-0a | GCGCAAGTCGACATTGTTCAG | |
| B10-1 RB-1a | ACGATGGACTCCAGTCCGGCCTGCCTTGCAGGTCGTACGCCATCAG | |
| B10-1 RB-2a | GTGCCCAGTACACGGCTCAGCGAC | |
| B13-1 LB-0a | CCGCCGTATGGATTCACCCGCAGCA | GS13-1植酸酶基因两翼序列扩增 |
| B13-1 LB-1a | ACGATGGACTCCAGTCCGGCCCGCTGAGCCGTGTACTGGGCACCAA | |
| B13-1 LB-2a | ACTGCTACAGGAGTTGCCGGTGGGG | |
| B13-1 RB-0a | CGCCCCACCGGCAACTCCTGTAGCA | |
| B13-1 RB-1a | ACGATGGACTCCAGTCCGGCCTTGCAGGTCGTACGCCATCAGTCCC | |
| B13-1 RB-2a | GCGACGGTTGCTGCGGGTGAATCCA |
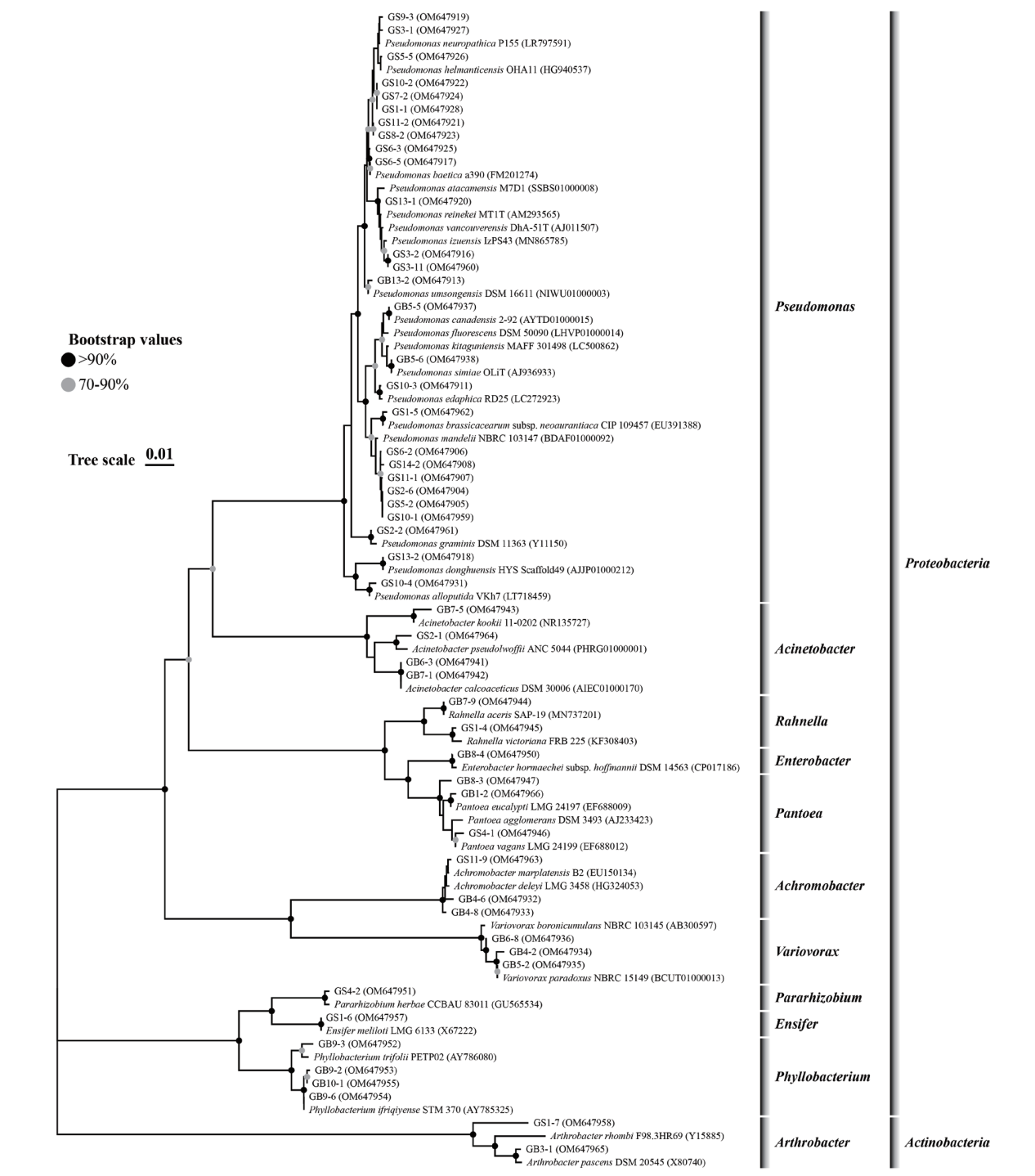
图1 基于16S rRNA基因序列构建的燕麦根际解植酸磷细菌系统发育树 有效序列长度为1 100 bp,邻接法构建发育树的自举值为1 000,括号中的序列号为菌株的GenBank登录号
Fig. 1 Phylogenetic tree of culturable bacteria from rhizosphere of Avena sativa and their closest relatives based on 16S rRNA gene sequences The effective sequence length was 1 100 bp, the bootstrap values were 1 000 replications in the Neighbour-Joining tree, and the serial number in parentheses denotes the GenBank accession number of the strains
| 菌株编号Strain code | 相似序列 Closest sequence | 相似度 Similarity/% | 植酸酶活性 Phytase activity/(U·mL-1) | 植酸酶基因扩增 Phytase gene presence | |
|---|---|---|---|---|---|
| BPPhy | HAPhy | ||||
| GB5-5 | Pseudomonas canadensis 2-92 | 100.00 | 0.103±0.007f | ﹣ | ﹣ |
| GB5-6 | Pseudomonas simiae OLiT | 100.00 | 0.119±0.005ef | ﹣ | ﹣ |
| GB13-2 | Pseudomonas umsongensis DSM 16611 | 99.78 | 0.149±0.021bc | ﹣ | ﹣ |
| GS1-5 | Pseudomonas brassicacearum subsp. neoaurantiaca CIP 109457 | 99.85 | 0.129±0.007cde | ﹣ | ﹣ |
| GS2-2 | Pseudomonas graminis DSM 11363 | 99.78 | 0.124±0.006def | ﹣ | ﹣ |
| GS3-1 | Pseudomonas neuropathica P155 | 99.84 | 0.189±0.025a | ﹣ | ﹣ |
| GS3-2 | Pseudomonas izuensis IzPS43 | 99.78 | 0.146±0.012bcd | ﹣ | ﹣ |
| GS5-5 | Pseudomonas helmanticensis OHA11 | 99.93 | 0.129±0.003cde | ﹣ | ﹣ |
| GS6-5 | Pseudomonas baeticaa390 | 100.00 | 0.161±0.012b | ﹢ | ﹣ |
| GS10-1 | Pseudomonas mandelii NBRC 103147 | 99.71 | 0.139±0.011bcde | ﹢ | ﹣ |
| GS10-3 | Pseudomonas edaphica RD25 | 99.85 | 0.137±0.016cde | ﹣ | ﹣ |
| GS10-4 | Pseudomonas alloputida VKh7 | 99.63 | 0.123±0.006def | ﹣ | ﹣ |
| GS13-1 | Pseudomonas reinekei MT1T | 99.78 | 0.141±0.005bcde | ﹢ | ﹣ |
| GS13-2 | Pseudomonas donghuensis HYS Scaffold49 | 100.00 | 0.129±0.004cde | ﹣ | ﹣ |
表2 假单胞菌属不同种菌株特性
Table 2 Characteristics of different species of Pseudomonas sp.
| 菌株编号Strain code | 相似序列 Closest sequence | 相似度 Similarity/% | 植酸酶活性 Phytase activity/(U·mL-1) | 植酸酶基因扩增 Phytase gene presence | |
|---|---|---|---|---|---|
| BPPhy | HAPhy | ||||
| GB5-5 | Pseudomonas canadensis 2-92 | 100.00 | 0.103±0.007f | ﹣ | ﹣ |
| GB5-6 | Pseudomonas simiae OLiT | 100.00 | 0.119±0.005ef | ﹣ | ﹣ |
| GB13-2 | Pseudomonas umsongensis DSM 16611 | 99.78 | 0.149±0.021bc | ﹣ | ﹣ |
| GS1-5 | Pseudomonas brassicacearum subsp. neoaurantiaca CIP 109457 | 99.85 | 0.129±0.007cde | ﹣ | ﹣ |
| GS2-2 | Pseudomonas graminis DSM 11363 | 99.78 | 0.124±0.006def | ﹣ | ﹣ |
| GS3-1 | Pseudomonas neuropathica P155 | 99.84 | 0.189±0.025a | ﹣ | ﹣ |
| GS3-2 | Pseudomonas izuensis IzPS43 | 99.78 | 0.146±0.012bcd | ﹣ | ﹣ |
| GS5-5 | Pseudomonas helmanticensis OHA11 | 99.93 | 0.129±0.003cde | ﹣ | ﹣ |
| GS6-5 | Pseudomonas baeticaa390 | 100.00 | 0.161±0.012b | ﹢ | ﹣ |
| GS10-1 | Pseudomonas mandelii NBRC 103147 | 99.71 | 0.139±0.011bcde | ﹢ | ﹣ |
| GS10-3 | Pseudomonas edaphica RD25 | 99.85 | 0.137±0.016cde | ﹣ | ﹣ |
| GS10-4 | Pseudomonas alloputida VKh7 | 99.63 | 0.123±0.006def | ﹣ | ﹣ |
| GS13-1 | Pseudomonas reinekei MT1T | 99.78 | 0.141±0.005bcde | ﹢ | ﹣ |
| GS13-2 | Pseudomonas donghuensis HYS Scaffold49 | 100.00 | 0.129±0.004cde | ﹣ | ﹣ |
| 菌株编号Strain code | 有机磷 Organic phosphorus | 无机磷 Inorganic phosphorus | IAA分泌量 IAA production/ (μg·mL-1) | 固氮酶活性 Nitrogenase activity/[nmol(C2H4)/(h·mL)] | 拮抗病原菌活性 Antifungal activity | |||
|---|---|---|---|---|---|---|---|---|
| pH | 溶磷量 P-solubilization/ (μg·mL-1) | pH | 溶磷量 P-solubilization/ (μg·mL-1) | 立枯丝核菌Rhizoctonia solani | 尖刀镰孢菌Fusarium oxysporum | |||
| GB5-5 | 4.28 | 460.73±40.36e | 5.17 | 412.29±39.70b | 0.24±0.05f | 51.29±2.56g | ﹢ | ﹣ |
| GB5-6 | 3.77 | 547.99±9.02a | 5.31 | 338.48±6.28c | ﹣ | 48.37±3.56g | ﹣ | ﹣ |
| GB13-2 | 3.65 | 535.06±11.82ab | 5.98 | 370.34±4.11bc | 0.41±0.11d | 68.44±1.90f | ﹣ | ﹣ |
| GS1-5 | 3.43 | 466.39±6.40e | 5.15 | 292.57±17.41d | 0.54±0.09d | 89.57±8.06abc | ﹣ | ﹣ |
| GS2-2 | 3.53 | 496.05±2.30cd | 5.26 | 418.78±68.58bc | 0.97±0.17c | 80.98±3.34cd | ﹢ | ﹣ |
| GS3-1 | 3.68 | 524.44±1.63ab | 5.96 | 264.82±1.99d | ﹣ | 85.50±4.47bcd | ﹢ | ﹣ |
| GS3-2 | 3.23 | 522.83±4.20abc | 5.43 | 419.14±7.41b | ﹣ | 69.51±6.06ef | ﹣ | ﹣ |
| GS5-5 | 3.80 | 474.35±14.09de | 6.02 | 372.63±14.73bc | 0.81±0.08c | 92.40±3.53ab | ﹣ | ﹣ |
| GS6-5 | 3.31 | 510.25±19.59bc | 5.09 | 468.66±60.61a | ﹣ | 84.90±3.79bcd | ﹣ | ﹢ |
| GS10-1 | 2.97 | 542.33±9.51a | 5.07 | 391.02±11.87b | 1.78±0.05a | 90.39±5.97abc | ﹢ | ﹢ |
| GS10-3 | 3.56 | 425.29±13.31f | 5.19 | 397.51±17.99b | ﹣ | 82.66±1.85bcd | ﹢ | ﹢ |
| GS10-4 | 3.52 | 465.23±12.00e | 5.48 | 380.92±5.55bc | 0.36±0.06e | 80.92±5.55cd | ﹢ | ﹣ |
| GS13-1 | 3.65 | 478.05±24.76de | 5.35 | 396.06±7.41b | ﹣ | 77.94±4.89de | ﹣ | ﹣ |
| GS13-2 | 3.88 | 480.01±5.74de | 5.28 | 421.66±9.98b | 1.27±0.52 bc | 97.47±10.12a | ﹢ | ﹣ |
表3 菌株促生特性分析
Table 3 Plant growth promoting characteristics of strains
| 菌株编号Strain code | 有机磷 Organic phosphorus | 无机磷 Inorganic phosphorus | IAA分泌量 IAA production/ (μg·mL-1) | 固氮酶活性 Nitrogenase activity/[nmol(C2H4)/(h·mL)] | 拮抗病原菌活性 Antifungal activity | |||
|---|---|---|---|---|---|---|---|---|
| pH | 溶磷量 P-solubilization/ (μg·mL-1) | pH | 溶磷量 P-solubilization/ (μg·mL-1) | 立枯丝核菌Rhizoctonia solani | 尖刀镰孢菌Fusarium oxysporum | |||
| GB5-5 | 4.28 | 460.73±40.36e | 5.17 | 412.29±39.70b | 0.24±0.05f | 51.29±2.56g | ﹢ | ﹣ |
| GB5-6 | 3.77 | 547.99±9.02a | 5.31 | 338.48±6.28c | ﹣ | 48.37±3.56g | ﹣ | ﹣ |
| GB13-2 | 3.65 | 535.06±11.82ab | 5.98 | 370.34±4.11bc | 0.41±0.11d | 68.44±1.90f | ﹣ | ﹣ |
| GS1-5 | 3.43 | 466.39±6.40e | 5.15 | 292.57±17.41d | 0.54±0.09d | 89.57±8.06abc | ﹣ | ﹣ |
| GS2-2 | 3.53 | 496.05±2.30cd | 5.26 | 418.78±68.58bc | 0.97±0.17c | 80.98±3.34cd | ﹢ | ﹣ |
| GS3-1 | 3.68 | 524.44±1.63ab | 5.96 | 264.82±1.99d | ﹣ | 85.50±4.47bcd | ﹢ | ﹣ |
| GS3-2 | 3.23 | 522.83±4.20abc | 5.43 | 419.14±7.41b | ﹣ | 69.51±6.06ef | ﹣ | ﹣ |
| GS5-5 | 3.80 | 474.35±14.09de | 6.02 | 372.63±14.73bc | 0.81±0.08c | 92.40±3.53ab | ﹣ | ﹣ |
| GS6-5 | 3.31 | 510.25±19.59bc | 5.09 | 468.66±60.61a | ﹣ | 84.90±3.79bcd | ﹣ | ﹢ |
| GS10-1 | 2.97 | 542.33±9.51a | 5.07 | 391.02±11.87b | 1.78±0.05a | 90.39±5.97abc | ﹢ | ﹢ |
| GS10-3 | 3.56 | 425.29±13.31f | 5.19 | 397.51±17.99b | ﹣ | 82.66±1.85bcd | ﹢ | ﹢ |
| GS10-4 | 3.52 | 465.23±12.00e | 5.48 | 380.92±5.55bc | 0.36±0.06e | 80.92±5.55cd | ﹢ | ﹣ |
| GS13-1 | 3.65 | 478.05±24.76de | 5.35 | 396.06±7.41b | ﹣ | 77.94±4.89de | ﹣ | ﹣ |
| GS13-2 | 3.88 | 480.01±5.74de | 5.28 | 421.66±9.98b | 1.27±0.52 bc | 97.47±10.12a | ﹢ | ﹣ |
| 基因编号 Gene code | 酶家族 Enzyme family | 相对分子质量 Molecular weight/kD | 理论等电点 Theoretical pI | 最相似菌株(UniProt ID) Most similar strain | 相似度 Similarity/% | GenBank登录号 GenBank accession No. |
|---|---|---|---|---|---|---|
| PHY65 | 3-phytase(EC 3.1.3.8) | 69.6 | 4.88 | Pseudomonas mucidolens (A0A1H2MGK9) | 78.3 | OM935858 |
| PHY101 | 3-phytase(EC 3.1.3.8) | 70.1 | 4.86 | Pseudomonas fluorescens (Q4KAB7) | 79.4 | OM935859 |
| PHY131 | 3-phytase(EC 3.1.3.8) | 69.8 | 4.84 | Pseudomonas fluorescens (Q4KAB7) | 78.1 | OM935857 |
表4 植酸酶氨基酸序列的基本性质
Table 4 Basic physicochemical properties of amino acid sequences of phytase
| 基因编号 Gene code | 酶家族 Enzyme family | 相对分子质量 Molecular weight/kD | 理论等电点 Theoretical pI | 最相似菌株(UniProt ID) Most similar strain | 相似度 Similarity/% | GenBank登录号 GenBank accession No. |
|---|---|---|---|---|---|---|
| PHY65 | 3-phytase(EC 3.1.3.8) | 69.6 | 4.88 | Pseudomonas mucidolens (A0A1H2MGK9) | 78.3 | OM935858 |
| PHY101 | 3-phytase(EC 3.1.3.8) | 70.1 | 4.86 | Pseudomonas fluorescens (Q4KAB7) | 79.4 | OM935859 |
| PHY131 | 3-phytase(EC 3.1.3.8) | 69.8 | 4.84 | Pseudomonas fluorescens (Q4KAB7) | 78.1 | OM935857 |
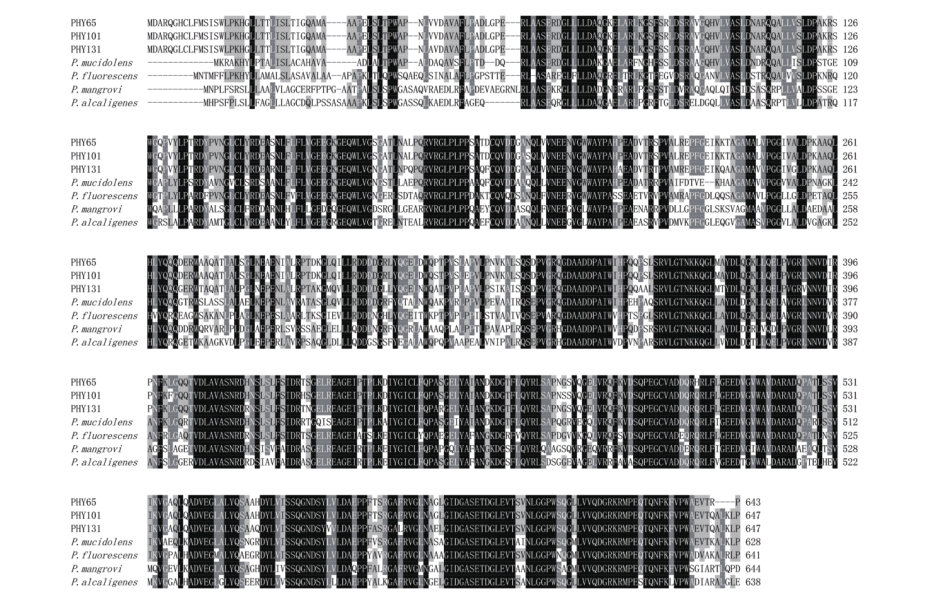
图3 PHY65、PHY101和PHY131的氨基酸多序列比对 阴影部分表示序列的相似程度,黑色表示序列完全相同,灰色表示序列高度相似。序列均来自GenBank,由上到下依次为:P. baetica PHY65(OM935858); P. mandelii PHY101(OM935859); P. reinekei PHY131(OM935857); P. mucidolens(WP084377901.1); P. fluorescens(WP014338283.1); P. alcaligenes(WP021702177.1); P. mangrovi(WP108107190.1)
Fig. 3 Amino acid multiple sequence alignment of PHY65, PHY101 and PHY131 The shaded part represents the degree of similarity of the sequences, black indicates that the sequences have completely identical and gray indicates that the sequences have highly similar. The sequences used in the figure are all from GenBank, and the access numbers from top to bottom are: P. baetica PHY65(OM935858); P. mandelii PHY101(OM935859); P. reinekei PHY131(OM935857); P. mucidolens(WP084377901.1); P. fluorescens(WP014338283.1); P. alcaligenes(WP021702177.1); P. mangrovi(WP108107190.1)
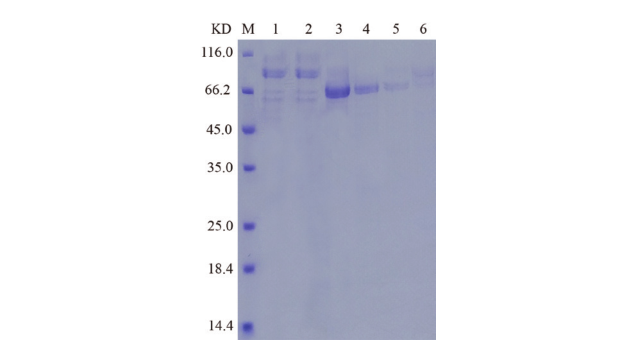
图4 蛋白表达鉴定SDS-PAGE分析 泳道M:蛋白标准品;泳道1:菌株分泌上清液;泳道2:过柱流出液;泳道3-6:过柱洗脱液
Fig. 4 SDS-PAGE analysis of protein expression M: Protein marker. Lane 1: The strain secretes supernatant. Lane 2: Effluent solution through column. Lane 3-6: Elution solution through column
| [1] | 王小春, 梁新强. 生态环境中植酸酶种类及来源分析[J]. 环境生态学, 2020, 2(4): 51-56. |
|
Wang XC, Liang XQ. Analysis of the types and sources of phytase in ecological environment[J]. Environ Ecol, 2020, 2(4): 51-56.
doi: 10.1890/1540-9295(2004)002[0051:PL]2.0.CO;2 URL |
|
| [2] |
丁锐, 陈旭辉, 李炳学. 植酸酶研究进展及土壤植酸酶应用展望[J]. 生物技术通报, 2019, 35(7): 190-195.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-1027 |
|
Ding R, Chen XH, Li BX. Research advances on phytase and prospect of applying soil phytase[J]. Biotechnol Bull, 2019, 35(7): 190-195.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-1027 |
|
| [3] |
Janes-Bassett V, Blackwell MSA, Blair G, et al. A meta-analysis of phosphatase activity in agricultural settings in response to phosphorus deficiency[J]. Soil Biol Biochem, 2022, 165: 108537.
doi: 10.1016/j.soilbio.2021.108537 URL |
| [4] |
Ushasree MV, Shyam K, et al. Microbial phytase: impact of advances in genetic engineering in revolutionizing its properties and applications[J]. Bioresour Technol, 2017, 245(Pt B): 1790-1799.
doi: 10.1016/j.biortech.2017.05.060 URL |
| [5] |
Singh B, Boukhris I, Pragya, et al. Contribution of microbial phytases to the improvement of plant growth and nutrition: a review[J]. Pedosphere, 2020, 30(3): 295-313.
doi: 10.1016/S1002-0160(20)60010-8 URL |
| [6] |
Balaban NP, Suleimanova AD, et al. Microbial phytases and phytate: exploring opportunities for sustainable phosphorus management in agriculture[J]. Am J Mol Biol, 2017, 7(1): 11-29.
doi: 10.4236/ajmb.2017.71002 URL |
| [7] |
Azeem M, Riaz A, Chaudhary AN, et al. Microbial phytase activity and their role in organic P mineralization[J]. Arch Agron Soil Sci, 2015, 61(6): 751-766.
doi: 10.1080/03650340.2014.963796 URL |
| [8] | 高亚敏, 等. 高寒草甸蒿草、珠芽蓼根际优良植物根际促生菌的分离筛选及促生特性研究[J]. 草业学报, 2019, 28(11): 114-123. |
| Gao YM, et al. Isolation, screening, and growth-promoting characteristics of plant growth promoting rhizobacteria in the rhizosphere of Kobresia myosuroides and Polygonum viviparum in alpine meadow pasture[J]. Acta Prataculturae Sin, 2019, 28(11): 114-123. | |
| [9] |
Li MY, Wang JL, Yao T, et al. Isolation and characterization of cold-adapted PGPB and their effect on plant growth promotion[J]. J Microbiol Biotechnol, 2021, 31(9): 1218-1230.
doi: 10.4014/jmb.2105.05012 URL |
| [10] | 张建贵, 王理德, 等. 祁连山高寒草地不同退化程度植物群落结构与物种多样性研究[J]. 草业学报, 2019(5): 15-25. |
| Zhang JG, Wang LD, et al. Plant community structure and species diversity differences in alpine grassland in the Qilian Mountains with different levels of degradation[J]. Acta Prataculturae Sin, 2019(5): 15-25. | |
| [11] |
George TS, Giles CD, et al. Organic phosphorus in the terrestrial environment: a perspective on the state of the art and future priorities[J]. Plant Soil, 2018, 427(1/2): 191-208.
doi: 10.1007/s11104-017-3391-x URL |
| [12] | 李明源, 王继莲, 姚拓, 等. 祁连山高寒草地扁蓿豆和黄花棘豆耐冷PGPB的筛选及促生特性研究[J]. 农业生物技术学报, 2021, 29(11): 2074-2086. |
| Li MY, Wang JL, Yao T, et al. Screening and promoting effects of cold-adapted PGPB from Melissitus Ruthenica and Oxytropis ochrocephala grown in the alpine grassland of Qilian mountains[J]. J Agric Biotechnol, 2021, 29(11): 2074-2086. | |
| [13] |
Bashan Y, Kamnev AA, de-Bashan LE. Tricalcium phosphate is inappropriate as a universal selection factor for isolating and testing phosphate-solubilizing bacteria that enhance plant growth: a proposal for an alternative procedure[J]. Biol Fertil Soils, 2013, 49(4): 465-479.
doi: 10.1007/s00374-012-0737-7 URL |
| [14] |
Mehta S, Nautiyal CS. An efficient method for qualitative screening of phosphate-solubilizing bacteria[J]. Curr Microbiol, 2001, 43(1): 51-56.
pmid: 11375664 |
| [15] | 刘婷, 姚拓, 陈建纲, 等. 固相萃取-高效液相色谱法测定植物根际促生菌发酵产物中3种植物激素的含量[J]. 分析科学学报, 2017, 33(2): 201-206. |
| Liu T, Yao T, Chen JG, et al. Determination of plant hormones in bacterial fermentation products of plant growth promoting rhizobacteria by solid phase extraction-high performance liquid chromatography[J]. J Anal Sci, 2017, 33(2): 201-206. | |
| [16] | Menezes-Blackburn D, Inostroza NG, Gianfreda L, et al. Phytase-producing Bacillus sp. inoculation increases phosphorus availability in cattle manure[J]. J Soil Sci Plant Nutr, 2016: 55108246. |
| [17] |
Yao QM, Li Z, Song Y, et al. Community proteogenomics reveals the systemic impact of phosphorus availability on microbial functions in tropical soil[J]. Nat Ecol Evol, 2018, 2(3): 499-509.
doi: 10.1038/s41559-017-0463-5 pmid: 29358607 |
| [18] |
Rasul M, Yasmin S, Yahya M, et al. The wheat growth-promoting traits of Ochrobactrum and Pantoea species, responsible for solubilization of different P sources, are ensured by genes encoding enzymes of multiple P-releasing pathways[J]. Microbiol Res, 2021, 246: 126703.
doi: 10.1016/j.micres.2021.126703 URL |
| [19] |
Liu YG, Chen YL. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences[J]. BioTechniques, 2007, 43(5): 649-656.
doi: 10.2144/000112601 URL |
| [20] |
Gessler NN, Serdyuk EG, Isakova EP, et al. Phytases and the prospects for their application(review)[J]. Appl Biochem Microbiol, 2018, 54(4): 352-360.
doi: 10.1134/S0003683818040087 |
| [21] |
渠露露, 彭长连, 李淑彬. 一株溶植酸磷类芽孢杆菌的分离筛选及对水稻幼苗的促生作用[J]. 应用生态学报, 2020, 31(1): 326-332.
doi: 10.13287/j.1001-9332.202001.033 |
| Qu LL, Peng CL, Li SB. Isolation and screening of a phytate phosphate-solubilizing Paenibacillus sp. and its growthpromoting effect on rice seeding[J]. Chin J Appl Ecol, 2020, 31(1): 326-332. | |
| [22] |
Kumar V, Yadav AN, et al. Β-Propeller phytases: Diversity, catalytic attributes, current developments and potential biotechnological applications[J]. Int J Biol Macromol, 2017, 98: 595-609.
doi: S0141-8130(16)32536-3 pmid: 28174082 |
| [23] |
Sanguin H, Wilson NL, Kertesz MA. Assessment of functional diversity and structure of phytate-hydrolysing bacterial community in Lolium perenne rhizosphere[J]. Plant Soil, 2016, 401(1/2): 151-167.
doi: 10.1007/s11104-015-2512-7 URL |
| [24] |
Wang F, Kertesz MA, Feng G. Phosphorus forms affect the hyphosphere bacterial community involved in soil organic phosphorus turnover[J]. Mycorrhiza, 2019, 29(4): 351-362.
doi: 10.1007/s00572-019-00896-0 pmid: 31044298 |
| [25] |
Wan WJ, Hao XL, Xing YH, et al. Spatial differences in soil microbial diversity caused by pH-driven organic phosphorus mineralization[J]. Land Degrad Dev, 2021, 32(2): 766-776.
doi: 10.1002/ldr.v32.2 URL |
| [26] |
杨婉秋, 敬洁, 朱灵, 等. 川西北高寒草甸植物根际促生菌筛选及其特性研究[J]. 草地学报, 2021, 29(6): 1174-1182.
doi: 10.11733/j.issn.1007-0435.2021.06.006 |
| Yang WQ, Jing J, Zhu L, et al. Screening and characteristics of plant growth-promoting rhizobacteria from alpine meadow plants in northwestern Sichuan[J]. Acta Agrestia Sin, 2021, 29(6): 1174-1182. | |
| [27] | 林启美, 赵小蓉, 孙焱鑫, 等. 四种不同生态系统的土壤解磷细菌数量及种群分布[J]. 土壤与环境, 2000, 9(1): 34-37. |
| Lin QM, Zhao XR, Sun YX, et al. Community characters of soil phosphobacteria in four ecosystems[J]. Soil Environ Sci, 2000, 9(1): 34-37. | |
| [28] | 周亚男, 韩小斌, 等. 烟草根际可培养微生物多样性及防病促生菌的筛选[J]. 微生物学通报, 2021, 48(12): 4649-4663. |
| Zhou YN, Han XB, et al. The culturable microbial diversity in tobacco rhizosphere and their plant growth-promoting and biocontrol properties[J]. Microbiol China, 2021, 48(12): 4649-4663. | |
| [29] |
Giles CD, George TS, Brown LK, et al. Does the combination of citrate and phytase exudation in Nicotiana tabacum promote the acquisition of endogenous soil organic phosphorus?[J]. Plant Soil, 2017, 412(1/2): 43-59.
doi: 10.1007/s11104-016-2884-3 URL |
| [30] |
Jang WJ, Lee JM, Park HD, et al. N-terminal domain of the beta-propeller phytase of Pseudomonas sp. FB15 plays a role for retention of low-temperature activity and catalytic efficiency[J]. Enzyme Microb Technol, 2018, 117: 84-90.
doi: 10.1016/j.enzmictec.2018.06.008 URL |
| [31] |
Jang WJ, Lee JM, Tawheed Hasan M, et al. Fusion of the N-terminal domain of Pseudomonas sp. phytase with Bacillus sp. phytase and its effects on optimal temperature and catalytic efficiency[J]. Enzyme Microb Technol, 2019, 126: 69-76.
doi: 10.1016/j.enzmictec.2019.04.002 URL |
| [1] | 张岳一, 兰社益, 裴海闰, 封棣. 多菌种联用发酵燕麦麸皮工艺优化及发用功效评价[J]. 生物技术通报, 2023, 39(9): 58-70. |
| [2] | 王琪媛, 王甲辰, 叶磊, 姜帆. 含ACC脱氨酶的根际细菌提高植物抗盐性的研究进展[J]. 生物技术通报, 2021, 37(2): 174-186. |
| [3] | 梁烨, 何楚婷, 杨悦, 张玉芬, 姜帆. 碱胁迫条件下含ACC脱氨酶的根际细菌对大豆生长的影响[J]. 生物技术通报, 2020, 36(9): 100-108. |
| [4] | 丁锐, 陈旭辉, 李炳学. 植酸酶研究进展及土壤植酸酶应用展望[J]. 生物技术通报, 2019, 35(7): 190-195. |
| [5] | 杨晓虹, 安江红, 韩冰, 周海涛, 杨才. 遮光模拟短日照对春夏播燕麦穗发育的影响[J]. 生物技术通报, 2018, 34(3): 93-97. |
| [6] | 伍甜,刘莹,余超,边强,田芳,黄琼,陈华民,何晨阳. 燕麦嗜酸菌鞭毛素的提纯及其活性检测[J]. 生物技术通报, 2013, 0(7): 78-81. |
| [7] | . 信息交流[J]. , 2007, 0(04): 9-63. |
| [8] | 张建社;刘明志;刘臻;. 植酸酶基因工程菌构建及其生产应用中问题分析[J]. , 2006, 0(05): 22-25. |
| [9] | . 农药[J]. , 1985, 0(09): 72-74. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||