








生物技术通报 ›› 2024, Vol. 40 ›› Issue (3): 202-214.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0982
收稿日期:2023-10-19
出版日期:2024-03-26
发布日期:2024-04-08
通讯作者:
刘新龙,男,博士,研究员,研究方向:甘蔗分子育种;E-mail: lxlgood868@163.com作者简介:田春艳,女,硕士,助理研究员,研究方向:甘蔗分子育种;E-mail: tianchy89@126.com
基金资助:
TIAN Chun-yan( ), LI Xu-juan, LI Chun-jia, MAO Jun, LIU Xin-long(
), LI Xu-juan, LI Chun-jia, MAO Jun, LIU Xin-long( )
)
Received:2023-10-19
Published:2024-03-26
Online:2024-04-08
摘要:
【目的】 为解析甘蔗基因组的密码子使用特征,提高异源基因在甘蔗中的表达效率。【方法】 以已发布的甘蔗属种(热带种LA-purple、割手密NP-X和AP85-441)及其近缘属种蔗茅(Yunnan2009-3)基因组为数据,利用Python、CodonW1.4.2进行密码子偏好性分析,同时通过中性绘图、ENC-plot、PR2-plot等分析探讨密码子偏好性形成的影响因素,并结合转录组测序数据分析密码子偏好性参数与基因表达水平的相关性。最后,基于RSCU均值与7个主要模式生物种(玉米、高粱、水稻、拟南芥、烟草、大肠杆菌、酿酒酵母)的密码子使用模式进行比较分析。【结果】 显示热带种、割手密和蔗茅的基因组都富含GC,平均GC含量为56.3%,且GC3>GC1>GC2, 倾向于使用以G/C结尾的密码子, 平均ENC值为48.45,偏好性较低。中性绘图、ENC-plot和PR2-plot分析表明它们的密码子偏好性受到自然选择、突变压力等多种因素的共同影响,其中自然选择占主导作用。相关性分析表明密码子偏好性参数与基因实际的转录表达水平存在显著相关性,但相关性不强。根据RSCU和∆RSCU值,确定了13个最优密码子,均以C或G结尾,密码子使用特性在全基因组和染色体组水平上无差异。通过比较发现,甘蔗的核苷酸组成及密码子偏好性与玉米、高粱和水稻较为相似,而与拟南芥、烟草、大肠杆菌和酵母具有显著差异。【结论】 甘蔗热带种、割手密和蔗茅的密码子偏好性高度相似,其形成受自然选择和突变因素的影响。此外,对甘蔗优异基因功能异源验证时可优先选择玉米、水稻和高粱作为异源表达系统。
田春艳, 李旭娟, 李纯佳, 毛钧, 刘新龙. 甘蔗属种及其近缘属种蔗茅的全基因组密码子偏好性分析[J]. 生物技术通报, 2024, 40(3): 202-214.
TIAN Chun-yan, LI Xu-juan, LI Chun-jia, MAO Jun, LIU Xin-long. Genome-wide Analysis of Codon Usage Bias in Saccharum Species and Its Phylogenetically Related Species Erianthus fulvus[J]. Biotechnology Bulletin, 2024, 40(3): 202-214.
| 核苷酸组成和ENC Nucleotide composition and ENC | 材料 Materials | 平均值 Mean | |||
|---|---|---|---|---|---|
| LA-purple | NP-X | AP85-441 | Yunnan2009-3 | ||
| A3s/% | 15.61±8.47 | 15.65±8.14 | 14.91±8.01 | 16.41±8.38 | 15.65 |
| T3s/% | 20.19±11.13 | 19.95±10.52 | 19.00±10.41 | 21.14±10.95 | 20.07 |
| G3s/% | 31.24±8.23 | 31.11±7.78 | 31.95±7.95 | 30.42±7.84 | 31.18 |
| C3s/% | 32.97±12.91 | 33.28±12.31 | 34.14±12.13 | 32.04±12.66 | 33.11 |
| GC/% | 56.21±0.10 | 56.42±0.09 | 57.53±0.09 | 55.02±0.09 | 56.30 |
| GC1/% | 58.39±7.29 | 58.53±6.76 | 59.29±6.80 | 57.67±6.86 | 58.47 |
| GC2/% | 45.73±7.79 | 46.05±7.34 | 46.87±7.57 | 44.67±7.06 | 45.83 |
| GC3/% | 64.21±18.91 | 64.39±18.02 | 66.09±17.78 | 62.46±18.68 | 64.29 |
| GC3s/% | 62.89±0.20 | 63.10±0.19 | 64.91±0.18 | 61.06±0.19 | 62.99 |
| ENC | 47.97±9.11 | 48.74±8.93 | 48.19±9.26 | 48.89±8.82 | 48.45 |
表1 甘蔗属种及其近缘属种蔗茅基因组CDS序列的核苷酸组成和ENC
Table 1 Nucleotide composition and ENC of CDS sequences in Saccharum species and its phylogenetically related species E. fulvus
| 核苷酸组成和ENC Nucleotide composition and ENC | 材料 Materials | 平均值 Mean | |||
|---|---|---|---|---|---|
| LA-purple | NP-X | AP85-441 | Yunnan2009-3 | ||
| A3s/% | 15.61±8.47 | 15.65±8.14 | 14.91±8.01 | 16.41±8.38 | 15.65 |
| T3s/% | 20.19±11.13 | 19.95±10.52 | 19.00±10.41 | 21.14±10.95 | 20.07 |
| G3s/% | 31.24±8.23 | 31.11±7.78 | 31.95±7.95 | 30.42±7.84 | 31.18 |
| C3s/% | 32.97±12.91 | 33.28±12.31 | 34.14±12.13 | 32.04±12.66 | 33.11 |
| GC/% | 56.21±0.10 | 56.42±0.09 | 57.53±0.09 | 55.02±0.09 | 56.30 |
| GC1/% | 58.39±7.29 | 58.53±6.76 | 59.29±6.80 | 57.67±6.86 | 58.47 |
| GC2/% | 45.73±7.79 | 46.05±7.34 | 46.87±7.57 | 44.67±7.06 | 45.83 |
| GC3/% | 64.21±18.91 | 64.39±18.02 | 66.09±17.78 | 62.46±18.68 | 64.29 |
| GC3s/% | 62.89±0.20 | 63.10±0.19 | 64.91±0.18 | 61.06±0.19 | 62.99 |
| ENC | 47.97±9.11 | 48.74±8.93 | 48.19±9.26 | 48.89±8.82 | 48.45 |
| ENC值 ENC value | LA-purple | NP-X | AP85-441 | Yunnan2009-3 | ||||
|---|---|---|---|---|---|---|---|---|
| 数目 | 百分比/% | 数目 | 百分比/% | 数目 | 百分比/% | 数目 | 百分比/% | |
| ENC≤35 | 34 381 | 14.15 | 15 880 | 12.10 | 8 642 | 14.23 | 4 290 | 12.05 |
| 35<ENC≤45 | 46 920 | 19.32 | 24 450 | 18.64 | 11 548 | 19.07 | 5 955 | 16.72 |
| ENC˃45 | 161 603 | 66.53 | 90 849 | 69.26 | 40 414 | 66.70 | 25 371 | 71.23 |
表2 ENC频数分布及百分比
Table 2 Frequency distribution and percentage of ENC
| ENC值 ENC value | LA-purple | NP-X | AP85-441 | Yunnan2009-3 | ||||
|---|---|---|---|---|---|---|---|---|
| 数目 | 百分比/% | 数目 | 百分比/% | 数目 | 百分比/% | 数目 | 百分比/% | |
| ENC≤35 | 34 381 | 14.15 | 15 880 | 12.10 | 8 642 | 14.23 | 4 290 | 12.05 |
| 35<ENC≤45 | 46 920 | 19.32 | 24 450 | 18.64 | 11 548 | 19.07 | 5 955 | 16.72 |
| ENC˃45 | 161 603 | 66.53 | 90 849 | 69.26 | 40 414 | 66.70 | 25 371 | 71.23 |
| 氨基酸 Amino acid | 密码子 Codon | LA-purple | NP-X | AP85-441 | Yunnan2009-3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GW | HEB | LEB | ∆RSCU | GW | HEB | LEB | ∆RSCU | GW | HEB | LEB | ∆RSCU | GW | HEB | LEB | ∆RSCU | |||||
| Phe | UUU | 0.76 | 0.05 | 0.95 | -0.90 | 0.73 | 0.06 | 0.95 | -0.89 | 0.69 | 0.19 | 0.93 | -0.74 | 0.78 | 0.06 | 0.96 | -0.90 | |||
| UUC** | 1.24 | 1.95 | 1.05 | 0.90 | 1.27 | 1.94 | 1.05 | 0.89 | 1.31 | 1.81 | 1.07 | 0.74 | 1.22 | 1.94 | 1.04 | 0.90 | ||||
| Leu | UUA | 0.39 | 0.01 | 0.53 | -0.52 | 0.37 | 0.01 | 0.52 | -0.51 | 0.33 | 0.04 | 0.51 | -0.47 | 0.41 | 0.01 | 0.53 | -0.52 | |||
| UUG | 0.94 | 0.14 | 1.11 | -0.97 | 0.92 | 0.16 | 1.11 | -0.95 | 0.86 | 0.08 | 1.10 | -1.02 | 0.97 | 0.15 | 1.12 | -0.97 | ||||
| CUU | 1.04 | 0.13 | 1.27 | -1.14 | 1.03 | 0.15 | 1.24 | -1.09 | 1.00 | 1.30 | 1.26 | 0.04 | 1.08 | 0.14 | 1.26 | -1.12 | ||||
| CUC** | 1.59 | 2.99 | 1.20 | 1.79 | 1.60 | 2.95 | 1.21 | 1.74 | 1.72 | 2.09 | 1.23 | 0.86 | 1.51 | 2.94 | 1.19 | 1.75 | ||||
| CUA | 0.50 | 0.07 | 0.66 | -0.59 | 0.50 | 0.08 | 0.67 | -0.59 | 0.46 | 0.88 | 0.65 | 0.23 | 0.54 | 0.08 | 0.67 | -0.59 | ||||
| CUG** | 1.54 | 2.65 | 1.24 | 1.41 | 1.58 | 2.65 | 1.24 | 1.41 | 1.64 | 1.61 | 1.26 | 0.35 | 1.49 | 2.67 | 1.22 | 1.45 | ||||
| Ile | AUU | 1.04 | 0.10 | 1.17 | -1.07 | 1.01 | 0.11 | 1.16 | -1.05 | 0.97 | 0.31 | 1.17 | -0.86 | 1.05 | 0.12 | 1.17 | -1.05 | |||
| AUC** | 1.34 | 2.81 | 1.04 | 1.77 | 1.35 | 2.78 | 1.04 | 1.74 | 1.43 | 2.48 | 1.05 | 1.43 | 1.29 | 2.78 | 1.03 | 1.75 | ||||
| AUA | 0.63 | 0.09 | 0.78 | -0.69 | 0.64 | 0.10 | 0.79 | -0.69 | 0.60 | 0.21 | 0.78 | -0.57 | 0.66 | 0.10 | 0.80 | -0.70 | ||||
| Val | GUU | 0.95 | 0.09 | 1.23 | -1.14 | 0.94 | 0.1 | 1.21 | -1.11 | 0.90 | 0.78 | 1.22 | -0.44 | 1.00 | 0.09 | 1.23 | -1.14 | |||
| GUC | 1.15 | 1.69 | 0.97 | 0.72 | 1.17 | 1.69 | 0.99 | 0.70 | 1.22 | 1.48 | 0.99 | 0.49 | 1.12 | 1.67 | 0.97 | 0.70 | ||||
| GUA | 0.41 | 0.04 | 0.58 | -0.54 | 0.41 | 0.05 | 0.58 | -0.53 | 0.38 | 0.53 | 0.57 | -0.04 | 0.44 | 0.05 | 0.59 | -0.54 | ||||
| GUG** | 1.48 | 2.18 | 1.22 | 0.96 | 1.48 | 2.17 | 1.22 | 0.95 | 1.50 | 1.21 | 1.01 | 0.20 | 1.44 | 2.19 | 1.21 | 0.98 | ||||
| Ser | UCU | 1.03 | 0.15 | 1.17 | -1.02 | 1.00 | 0.16 | 1.16 | -1.00 | 0.96 | 0.58 | 1.14 | -0.56 | 1.06 | 0.16 | 1.17 | -1.01 | |||
| UCC** | 1.25 | 2.24 | 1.06 | 1.18 | 1.24 | 2.18 | 1.07 | 1.11 | 1.32 | 1.85 | 1.08 | 0.77 | 1.19 | 2.17 | 1.03 | 1.14 | ||||
| UCA | 0.99 | 0.13 | 1.18 | -1.05 | 0.99 | 0.14 | 1.17 | -1.03 | 0.94 | 0.33 | 1.16 | -0.83 | 1.04 | 0.14 | 1.20 | -1.06 | ||||
| UCG | 0.80 | 1.65 | 0.68 | 0.97 | 0.81 | 1.64 | 0.69 | 0.95 | 0.87 | 1.21 | 0.70 | 0.51 | 0.76 | 1.66 | 0.68 | 0.98 | ||||
| AGU | 0.72 | 0.07 | 0.85 | -0.78 | 0.70 | 0.08 | 0.85 | -0.77 | 0.64 | 0.30 | 0.84 | -0.54 | 0.75 | 0.08 | 0.87 | -0.79 | ||||
| AGC** | 1.22 | 1.76 | 1.05 | 0.71 | 1.25 | 1.80 | 1.07 | 0.73 | 1.27 | 1.73 | 1.07 | 0.66 | 1.20 | 1.79 | 1.05 | 0.74 | ||||
| Pro | CCU | 0.98 | 0.20 | 1.15 | -0.95 | 0.96 | 0.21 | 1.13 | -0.92 | 0.92 | 1.20 | 1.14 | 0.06 | 1.02 | 0.21 | 1.17 | -0.96 | |||
| CCC | 0.88 | 1.38 | 0.80 | 0.58 | 0.88 | 1.36 | 0.81 | 0.55 | 0.92 | 0.95 | 0.82 | 0.13 | 0.85 | 1.36 | 0.79 | 0.57 | ||||
| CCA | 1.03 | 0.20 | 1.20 | -1.00 | 1.03 | 0.22 | 1.20 | -0.98 | 0.96 | 0.70 | 1.18 | -0.48 | 1.08 | 0.21 | 1.22 | -1.01 | ||||
| CCG | 1.11 | 2.22 | 0.85 | 1.37 | 1.12 | 2.21 | 0.85 | 1.36 | 1.20 | 1.14 | 0.86 | 0.28 | 1.05 | 2.22 | 0.82 | 1.40 | ||||
| Thr | ACU | 0.95 | 0.10 | 1.13 | -1.03 | 0.91 | 0.11 | 1.12 | -1.01 | 0.87 | 0.38 | 1.12 | -0.74 | 0.97 | 0.11 | 1.14 | -1.03 | |||
| ACC | 1.20 | 1.91 | 1.00 | 0.91 | 1.20 | 1.87 | 1.00 | 0.87 | 1.26 | 1.72 | 1.01 | 0.71 | 1.17 | 1.87 | 0.98 | 0.89 | ||||
| ACA | 1.00 | 0.11 | 1.22 | -1.11 | 1.01 | 0.12 | 1.21 | -1.09 | 0.94 | 0.33 | 1.21 | -0.88 | 1.06 | 0.12 | 1.24 | -1.12 | ||||
| ACG | 0.85 | 1.88 | 0.65 | 1.23 | 0.88 | 1.89 | 0.67 | 1.22 | 0.93 | 1.58 | 0.67 | 0.91 | 0.81 | 1.90 | 0.65 | 1.25 | ||||
| Ala | GCU | 0.92 | 0.15 | 1.16 | -1.01 | 0.90 | 0.16 | 1.14 | -0.98 | 0.85 | 1.21 | 1.14 | 0.07 | 0.95 | 0.16 | 1.17 | -1.01 | |||
| GCC** | 1.27 | 1.91 | 1.02 | 0.89 | 1.28 | 1.90 | 1.03 | 0.87 | 1.33 | 1.14 | 1.03 | 0.11 | 1.24 | 1.88 | 1.02 | 0.86 | ||||
| GCA | 0.81 | 0.14 | 1.06 | -0.92 | 0.81 | 0.15 | 1.05 | -0.90 | 0.75 | 0.66 | 1.04 | -0.38 | 0.86 | 0.14 | 1.06 | -0.92 | ||||
| GCG | 1.00 | 1.80 | 0.76 | 1.04 | 1.01 | 1.79 | 0.78 | 1.01 | 1.07 | 1.00 | 0.79 | 0.21 | 0.95 | 1.81 | 0.75 | 1.06 | ||||
| Tyr | UAU | 0.81 | 0.05 | 1.00 | -0.95 | 0.77 | 0.05 | 1.01 | -0.96 | 0.72 | 0.2 | 0.99 | -0.79 | 0.83 | 0.05 | 1.02 | -0.97 | |||
| UAC** | 1.19 | 1.95 | 1.00 | 0.95 | 1.23 | 1.95 | 0.99 | 0.96 | 1.28 | 1.80 | 1.01 | 0.79 | 1.17 | 1.95 | 0.98 | 0.97 | ||||
| His | CAU | 0.93 | 0.12 | 1.09 | -0.97 | 0.90 | 0.13 | 1.08 | -0.95 | 0.84 | 0.95 | 1.07 | -0.12 | 0.96 | 0.13 | 1.10 | -0.97 | |||
| CAC | 1.07 | 1.88 | 0.91 | 0.97 | 1.10 | 1.87 | 0.92 | 0.95 | 1.16 | 1.05 | 0.93 | 0.12 | 1.04 | 1.87 | 0.90 | 0.97 | ||||
| Gln | CAA | 0.75 | 0.10 | 0.88 | -0.78 | 0.73 | 0.11 | 0.88 | -0.77 | 0.67 | 1.19 | 0.86 | 0.33 | 0.79 | 0.11 | 0.90 | -0.79 | |||
| CAG* | 1.25 | 1.90 | 1.12 | 0.78 | 1.27 | 1.89 | 1.12 | 0.77 | 1.33 | 0.81 | 1.14 | -0.33 | 1.21 | 1.89 | 1.10 | 0.79 | ||||
| Asn | AAU | 0.92 | 0.09 | 1.06 | -0.97 | 0.90 | 0.10 | 1.06 | -0.96 | 0.86 | 0.61 | 1.04 | -0.43 | 0.95 | 0.11 | 1.06 | -0.95 | |||
| AAC | 1.08 | 1.91 | 0.94 | 0.97 | 1.10 | 1.90 | 0.94 | 0.96 | 1.14 | 1.39 | 0.96 | 0.43 | 1.05 | 1.89 | 0.94 | 0.95 | ||||
| Lys | AAA | 0.65 | 0.09 | 0.79 | -0.70 | 0.66 | 0.09 | 0.80 | -0.71 | 0.62 | 0.25 | 0.79 | -0.54 | 0.68 | 0.09 | 0.80 | -0.71 | |||
| AAG** | 1.35 | 1.91 | 1.21 | 0.70 | 1.34 | 1.91 | 1.2 | 0.71 | 1.38 | 1.75 | 1.21 | 0.54 | 1.32 | 1.91 | 1.20 | 0.71 | ||||
| Asp | GAU | 0.96 | 0.12 | 1.15 | -1.03 | 0.94 | 0.12 | 1.14 | -1.02 | 0.89 | 0.77 | 1.14 | -0.37 | 0.99 | 0.12 | 1.15 | -1.03 | |||
| GAC | 1.04 | 1.88 | 0.85 | 1.03 | 1.06 | 1.88 | 0.86 | 1.02 | 1.11 | 1.23 | 0.86 | 0.37 | 1.01 | 1.88 | 0.85 | 1.03 | ||||
| Glu | GAA | 0.73 | 0.10 | 0.87 | -0.77 | 0.73 | 0.12 | 0.88 | -0.76 | 0.68 | 0.90 | 0.87 | 0.03 | 0.77 | 0.11 | 0.88 | -0.77 | |||
| GAG* | 1.27 | 1.90 | 1.13 | 0.77 | 1.27 | 1.88 | 1.12 | 0.76 | 1.32 | 1.10 | 1.13 | -0.03 | 1.23 | 1.89 | 1.12 | 0.77 | ||||
| Cys | UGU | 0.70 | 0.06 | 0.87 | -0.81 | 0.67 | 0.06 | 0.86 | -0.80 | 0.62 | 0.42 | 0.84 | -0.42 | 0.72 | 0.06 | 0.87 | -0.81 | |||
| UGC** | 1.30 | 1.94 | 1.13 | 0.81 | 1.33 | 1.94 | 1.14 | 0.80 | 1.38 | 1.58 | 1.16 | 0.42 | 1.28 | 1.94 | 1.13 | 0.81 | ||||
| Arg | CGU | 0.63 | 0.17 | 0.77 | -0.60 | 0.62 | 0.18 | 0.77 | -0.59 | 0.59 | 1.15 | 0.76 | 0.39 | 0.62 | 0.18 | 0.77 | -0.59 | |||
| CGC** | 1.42 | 2.86 | 1.10 | 1.76 | 1.43 | 2.79 | 1.11 | 1.68 | 1.53 | 1.92 | 1.11 | 0.81 | 1.35 | 2.78 | 1.10 | 1.68 | ||||
| CGA | 0.45 | 0.11 | 0.63 | -0.52 | 0.46 | 0.12 | 0.64 | -0.52 | 0.45 | 1.28 | 0.64 | 0.64 | 0.46 | 0.12 | 0.63 | -0.51 | ||||
| CGG | 1.06 | 1.82 | 0.94 | 0.88 | 1.09 | 1.82 | 0.96 | 0.86 | 1.17 | 1.44 | 0.98 | 0.46 | 1.02 | 1.84 | 0.94 | 0.90 | ||||
| AGA | 1.00 | 0.09 | 1.19 | -1.10 | 0.96 | 0.18 | 0.77 | -0.59 | 0.87 | 0.06 | 1.15 | -1.09 | 1.07 | 0.10 | 1.20 | -1.10 | ||||
| AGG | 1.44 | 0.95 | 1.38 | -0.43 | 1.45 | 2.79 | 1.11 | 1.68 | 1.39 | 0.15 | 1.35 | -1.20 | 1.48 | 0.98 | 1.37 | -0.39 | ||||
| Gly | GGU | 0.83 | 0.17 | 1.02 | -0.85 | 0.81 | 0.17 | 1.00 | -0.83 | 0.77 | 0.67 | 0.98 | -0.31 | 0.86 | 0.16 | 1.02 | -0.86 | |||
| GGC** | 1.52 | 2.67 | 1.10 | 1.57 | 1.53 | 2.65 | 1.13 | 1.52 | 1.59 | 1.57 | 1.13 | 0.44 | 1.47 | 2.65 | 1.10 | 1.55 | ||||
| GGA | 0.79 | 0.19 | 1.01 | -0.82 | 0.79 | 0.20 | 1.02 | -0.82 | 0.75 | 1.03 | 1.01 | 0.02 | 0.82 | 0.19 | 1.02 | -0.83 | ||||
| GGG | 0.86 | 0.98 | 0.87 | 0.11 | 0.87 | 0.98 | 0.86 | 0.12 | 0.89 | 0.73 | 0.88 | -0.15 | 0.86 | 0.99 | 0.86 | 0.13 | ||||
| Trp | UGG | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | |||
| Met | AUG | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | |||
| Ter | UAA | 0.93 | 0.38 | 0.74 | -0.36 | 0.66 | 0.42 | 0.73 | -0.31 | 0.66 | 0.54 | 0.73 | -0.19 | 0.67 | 0.38 | 0.73 | -0.35 | |||
| UAG | 0.93 | 0.90 | 0.90 | 0.00 | 0.93 | 0.92 | 0.91 | 0.01 | 0.92 | 0.50 | 0.89 | -0.39 | 0.94 | 0.91 | 0.91 | 0.00 | ||||
| UGA | 1.42 | 1.00 | 1.00 | 0.00 | 1.41 | 1.00 | 1.00 | 0.00 | 1.43 | 1.96 | 1.38 | 0.58 | 1.39 | 1.70 | 1.35 | 0.35 | ||||
表3 甘蔗属种及其近缘属种蔗茅的最优密码子分析
Table 3 Optimal codons analysis of Saccharum species and its phylogenetically related species E. fulvus
| 氨基酸 Amino acid | 密码子 Codon | LA-purple | NP-X | AP85-441 | Yunnan2009-3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GW | HEB | LEB | ∆RSCU | GW | HEB | LEB | ∆RSCU | GW | HEB | LEB | ∆RSCU | GW | HEB | LEB | ∆RSCU | |||||
| Phe | UUU | 0.76 | 0.05 | 0.95 | -0.90 | 0.73 | 0.06 | 0.95 | -0.89 | 0.69 | 0.19 | 0.93 | -0.74 | 0.78 | 0.06 | 0.96 | -0.90 | |||
| UUC** | 1.24 | 1.95 | 1.05 | 0.90 | 1.27 | 1.94 | 1.05 | 0.89 | 1.31 | 1.81 | 1.07 | 0.74 | 1.22 | 1.94 | 1.04 | 0.90 | ||||
| Leu | UUA | 0.39 | 0.01 | 0.53 | -0.52 | 0.37 | 0.01 | 0.52 | -0.51 | 0.33 | 0.04 | 0.51 | -0.47 | 0.41 | 0.01 | 0.53 | -0.52 | |||
| UUG | 0.94 | 0.14 | 1.11 | -0.97 | 0.92 | 0.16 | 1.11 | -0.95 | 0.86 | 0.08 | 1.10 | -1.02 | 0.97 | 0.15 | 1.12 | -0.97 | ||||
| CUU | 1.04 | 0.13 | 1.27 | -1.14 | 1.03 | 0.15 | 1.24 | -1.09 | 1.00 | 1.30 | 1.26 | 0.04 | 1.08 | 0.14 | 1.26 | -1.12 | ||||
| CUC** | 1.59 | 2.99 | 1.20 | 1.79 | 1.60 | 2.95 | 1.21 | 1.74 | 1.72 | 2.09 | 1.23 | 0.86 | 1.51 | 2.94 | 1.19 | 1.75 | ||||
| CUA | 0.50 | 0.07 | 0.66 | -0.59 | 0.50 | 0.08 | 0.67 | -0.59 | 0.46 | 0.88 | 0.65 | 0.23 | 0.54 | 0.08 | 0.67 | -0.59 | ||||
| CUG** | 1.54 | 2.65 | 1.24 | 1.41 | 1.58 | 2.65 | 1.24 | 1.41 | 1.64 | 1.61 | 1.26 | 0.35 | 1.49 | 2.67 | 1.22 | 1.45 | ||||
| Ile | AUU | 1.04 | 0.10 | 1.17 | -1.07 | 1.01 | 0.11 | 1.16 | -1.05 | 0.97 | 0.31 | 1.17 | -0.86 | 1.05 | 0.12 | 1.17 | -1.05 | |||
| AUC** | 1.34 | 2.81 | 1.04 | 1.77 | 1.35 | 2.78 | 1.04 | 1.74 | 1.43 | 2.48 | 1.05 | 1.43 | 1.29 | 2.78 | 1.03 | 1.75 | ||||
| AUA | 0.63 | 0.09 | 0.78 | -0.69 | 0.64 | 0.10 | 0.79 | -0.69 | 0.60 | 0.21 | 0.78 | -0.57 | 0.66 | 0.10 | 0.80 | -0.70 | ||||
| Val | GUU | 0.95 | 0.09 | 1.23 | -1.14 | 0.94 | 0.1 | 1.21 | -1.11 | 0.90 | 0.78 | 1.22 | -0.44 | 1.00 | 0.09 | 1.23 | -1.14 | |||
| GUC | 1.15 | 1.69 | 0.97 | 0.72 | 1.17 | 1.69 | 0.99 | 0.70 | 1.22 | 1.48 | 0.99 | 0.49 | 1.12 | 1.67 | 0.97 | 0.70 | ||||
| GUA | 0.41 | 0.04 | 0.58 | -0.54 | 0.41 | 0.05 | 0.58 | -0.53 | 0.38 | 0.53 | 0.57 | -0.04 | 0.44 | 0.05 | 0.59 | -0.54 | ||||
| GUG** | 1.48 | 2.18 | 1.22 | 0.96 | 1.48 | 2.17 | 1.22 | 0.95 | 1.50 | 1.21 | 1.01 | 0.20 | 1.44 | 2.19 | 1.21 | 0.98 | ||||
| Ser | UCU | 1.03 | 0.15 | 1.17 | -1.02 | 1.00 | 0.16 | 1.16 | -1.00 | 0.96 | 0.58 | 1.14 | -0.56 | 1.06 | 0.16 | 1.17 | -1.01 | |||
| UCC** | 1.25 | 2.24 | 1.06 | 1.18 | 1.24 | 2.18 | 1.07 | 1.11 | 1.32 | 1.85 | 1.08 | 0.77 | 1.19 | 2.17 | 1.03 | 1.14 | ||||
| UCA | 0.99 | 0.13 | 1.18 | -1.05 | 0.99 | 0.14 | 1.17 | -1.03 | 0.94 | 0.33 | 1.16 | -0.83 | 1.04 | 0.14 | 1.20 | -1.06 | ||||
| UCG | 0.80 | 1.65 | 0.68 | 0.97 | 0.81 | 1.64 | 0.69 | 0.95 | 0.87 | 1.21 | 0.70 | 0.51 | 0.76 | 1.66 | 0.68 | 0.98 | ||||
| AGU | 0.72 | 0.07 | 0.85 | -0.78 | 0.70 | 0.08 | 0.85 | -0.77 | 0.64 | 0.30 | 0.84 | -0.54 | 0.75 | 0.08 | 0.87 | -0.79 | ||||
| AGC** | 1.22 | 1.76 | 1.05 | 0.71 | 1.25 | 1.80 | 1.07 | 0.73 | 1.27 | 1.73 | 1.07 | 0.66 | 1.20 | 1.79 | 1.05 | 0.74 | ||||
| Pro | CCU | 0.98 | 0.20 | 1.15 | -0.95 | 0.96 | 0.21 | 1.13 | -0.92 | 0.92 | 1.20 | 1.14 | 0.06 | 1.02 | 0.21 | 1.17 | -0.96 | |||
| CCC | 0.88 | 1.38 | 0.80 | 0.58 | 0.88 | 1.36 | 0.81 | 0.55 | 0.92 | 0.95 | 0.82 | 0.13 | 0.85 | 1.36 | 0.79 | 0.57 | ||||
| CCA | 1.03 | 0.20 | 1.20 | -1.00 | 1.03 | 0.22 | 1.20 | -0.98 | 0.96 | 0.70 | 1.18 | -0.48 | 1.08 | 0.21 | 1.22 | -1.01 | ||||
| CCG | 1.11 | 2.22 | 0.85 | 1.37 | 1.12 | 2.21 | 0.85 | 1.36 | 1.20 | 1.14 | 0.86 | 0.28 | 1.05 | 2.22 | 0.82 | 1.40 | ||||
| Thr | ACU | 0.95 | 0.10 | 1.13 | -1.03 | 0.91 | 0.11 | 1.12 | -1.01 | 0.87 | 0.38 | 1.12 | -0.74 | 0.97 | 0.11 | 1.14 | -1.03 | |||
| ACC | 1.20 | 1.91 | 1.00 | 0.91 | 1.20 | 1.87 | 1.00 | 0.87 | 1.26 | 1.72 | 1.01 | 0.71 | 1.17 | 1.87 | 0.98 | 0.89 | ||||
| ACA | 1.00 | 0.11 | 1.22 | -1.11 | 1.01 | 0.12 | 1.21 | -1.09 | 0.94 | 0.33 | 1.21 | -0.88 | 1.06 | 0.12 | 1.24 | -1.12 | ||||
| ACG | 0.85 | 1.88 | 0.65 | 1.23 | 0.88 | 1.89 | 0.67 | 1.22 | 0.93 | 1.58 | 0.67 | 0.91 | 0.81 | 1.90 | 0.65 | 1.25 | ||||
| Ala | GCU | 0.92 | 0.15 | 1.16 | -1.01 | 0.90 | 0.16 | 1.14 | -0.98 | 0.85 | 1.21 | 1.14 | 0.07 | 0.95 | 0.16 | 1.17 | -1.01 | |||
| GCC** | 1.27 | 1.91 | 1.02 | 0.89 | 1.28 | 1.90 | 1.03 | 0.87 | 1.33 | 1.14 | 1.03 | 0.11 | 1.24 | 1.88 | 1.02 | 0.86 | ||||
| GCA | 0.81 | 0.14 | 1.06 | -0.92 | 0.81 | 0.15 | 1.05 | -0.90 | 0.75 | 0.66 | 1.04 | -0.38 | 0.86 | 0.14 | 1.06 | -0.92 | ||||
| GCG | 1.00 | 1.80 | 0.76 | 1.04 | 1.01 | 1.79 | 0.78 | 1.01 | 1.07 | 1.00 | 0.79 | 0.21 | 0.95 | 1.81 | 0.75 | 1.06 | ||||
| Tyr | UAU | 0.81 | 0.05 | 1.00 | -0.95 | 0.77 | 0.05 | 1.01 | -0.96 | 0.72 | 0.2 | 0.99 | -0.79 | 0.83 | 0.05 | 1.02 | -0.97 | |||
| UAC** | 1.19 | 1.95 | 1.00 | 0.95 | 1.23 | 1.95 | 0.99 | 0.96 | 1.28 | 1.80 | 1.01 | 0.79 | 1.17 | 1.95 | 0.98 | 0.97 | ||||
| His | CAU | 0.93 | 0.12 | 1.09 | -0.97 | 0.90 | 0.13 | 1.08 | -0.95 | 0.84 | 0.95 | 1.07 | -0.12 | 0.96 | 0.13 | 1.10 | -0.97 | |||
| CAC | 1.07 | 1.88 | 0.91 | 0.97 | 1.10 | 1.87 | 0.92 | 0.95 | 1.16 | 1.05 | 0.93 | 0.12 | 1.04 | 1.87 | 0.90 | 0.97 | ||||
| Gln | CAA | 0.75 | 0.10 | 0.88 | -0.78 | 0.73 | 0.11 | 0.88 | -0.77 | 0.67 | 1.19 | 0.86 | 0.33 | 0.79 | 0.11 | 0.90 | -0.79 | |||
| CAG* | 1.25 | 1.90 | 1.12 | 0.78 | 1.27 | 1.89 | 1.12 | 0.77 | 1.33 | 0.81 | 1.14 | -0.33 | 1.21 | 1.89 | 1.10 | 0.79 | ||||
| Asn | AAU | 0.92 | 0.09 | 1.06 | -0.97 | 0.90 | 0.10 | 1.06 | -0.96 | 0.86 | 0.61 | 1.04 | -0.43 | 0.95 | 0.11 | 1.06 | -0.95 | |||
| AAC | 1.08 | 1.91 | 0.94 | 0.97 | 1.10 | 1.90 | 0.94 | 0.96 | 1.14 | 1.39 | 0.96 | 0.43 | 1.05 | 1.89 | 0.94 | 0.95 | ||||
| Lys | AAA | 0.65 | 0.09 | 0.79 | -0.70 | 0.66 | 0.09 | 0.80 | -0.71 | 0.62 | 0.25 | 0.79 | -0.54 | 0.68 | 0.09 | 0.80 | -0.71 | |||
| AAG** | 1.35 | 1.91 | 1.21 | 0.70 | 1.34 | 1.91 | 1.2 | 0.71 | 1.38 | 1.75 | 1.21 | 0.54 | 1.32 | 1.91 | 1.20 | 0.71 | ||||
| Asp | GAU | 0.96 | 0.12 | 1.15 | -1.03 | 0.94 | 0.12 | 1.14 | -1.02 | 0.89 | 0.77 | 1.14 | -0.37 | 0.99 | 0.12 | 1.15 | -1.03 | |||
| GAC | 1.04 | 1.88 | 0.85 | 1.03 | 1.06 | 1.88 | 0.86 | 1.02 | 1.11 | 1.23 | 0.86 | 0.37 | 1.01 | 1.88 | 0.85 | 1.03 | ||||
| Glu | GAA | 0.73 | 0.10 | 0.87 | -0.77 | 0.73 | 0.12 | 0.88 | -0.76 | 0.68 | 0.90 | 0.87 | 0.03 | 0.77 | 0.11 | 0.88 | -0.77 | |||
| GAG* | 1.27 | 1.90 | 1.13 | 0.77 | 1.27 | 1.88 | 1.12 | 0.76 | 1.32 | 1.10 | 1.13 | -0.03 | 1.23 | 1.89 | 1.12 | 0.77 | ||||
| Cys | UGU | 0.70 | 0.06 | 0.87 | -0.81 | 0.67 | 0.06 | 0.86 | -0.80 | 0.62 | 0.42 | 0.84 | -0.42 | 0.72 | 0.06 | 0.87 | -0.81 | |||
| UGC** | 1.30 | 1.94 | 1.13 | 0.81 | 1.33 | 1.94 | 1.14 | 0.80 | 1.38 | 1.58 | 1.16 | 0.42 | 1.28 | 1.94 | 1.13 | 0.81 | ||||
| Arg | CGU | 0.63 | 0.17 | 0.77 | -0.60 | 0.62 | 0.18 | 0.77 | -0.59 | 0.59 | 1.15 | 0.76 | 0.39 | 0.62 | 0.18 | 0.77 | -0.59 | |||
| CGC** | 1.42 | 2.86 | 1.10 | 1.76 | 1.43 | 2.79 | 1.11 | 1.68 | 1.53 | 1.92 | 1.11 | 0.81 | 1.35 | 2.78 | 1.10 | 1.68 | ||||
| CGA | 0.45 | 0.11 | 0.63 | -0.52 | 0.46 | 0.12 | 0.64 | -0.52 | 0.45 | 1.28 | 0.64 | 0.64 | 0.46 | 0.12 | 0.63 | -0.51 | ||||
| CGG | 1.06 | 1.82 | 0.94 | 0.88 | 1.09 | 1.82 | 0.96 | 0.86 | 1.17 | 1.44 | 0.98 | 0.46 | 1.02 | 1.84 | 0.94 | 0.90 | ||||
| AGA | 1.00 | 0.09 | 1.19 | -1.10 | 0.96 | 0.18 | 0.77 | -0.59 | 0.87 | 0.06 | 1.15 | -1.09 | 1.07 | 0.10 | 1.20 | -1.10 | ||||
| AGG | 1.44 | 0.95 | 1.38 | -0.43 | 1.45 | 2.79 | 1.11 | 1.68 | 1.39 | 0.15 | 1.35 | -1.20 | 1.48 | 0.98 | 1.37 | -0.39 | ||||
| Gly | GGU | 0.83 | 0.17 | 1.02 | -0.85 | 0.81 | 0.17 | 1.00 | -0.83 | 0.77 | 0.67 | 0.98 | -0.31 | 0.86 | 0.16 | 1.02 | -0.86 | |||
| GGC** | 1.52 | 2.67 | 1.10 | 1.57 | 1.53 | 2.65 | 1.13 | 1.52 | 1.59 | 1.57 | 1.13 | 0.44 | 1.47 | 2.65 | 1.10 | 1.55 | ||||
| GGA | 0.79 | 0.19 | 1.01 | -0.82 | 0.79 | 0.20 | 1.02 | -0.82 | 0.75 | 1.03 | 1.01 | 0.02 | 0.82 | 0.19 | 1.02 | -0.83 | ||||
| GGG | 0.86 | 0.98 | 0.87 | 0.11 | 0.87 | 0.98 | 0.86 | 0.12 | 0.89 | 0.73 | 0.88 | -0.15 | 0.86 | 0.99 | 0.86 | 0.13 | ||||
| Trp | UGG | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | |||
| Met | AUG | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | |||
| Ter | UAA | 0.93 | 0.38 | 0.74 | -0.36 | 0.66 | 0.42 | 0.73 | -0.31 | 0.66 | 0.54 | 0.73 | -0.19 | 0.67 | 0.38 | 0.73 | -0.35 | |||
| UAG | 0.93 | 0.90 | 0.90 | 0.00 | 0.93 | 0.92 | 0.91 | 0.01 | 0.92 | 0.50 | 0.89 | -0.39 | 0.94 | 0.91 | 0.91 | 0.00 | ||||
| UGA | 1.42 | 1.00 | 1.00 | 0.00 | 1.41 | 1.00 | 1.00 | 0.00 | 1.43 | 1.96 | 1.38 | 0.58 | 1.39 | 1.70 | 1.35 | 0.35 | ||||
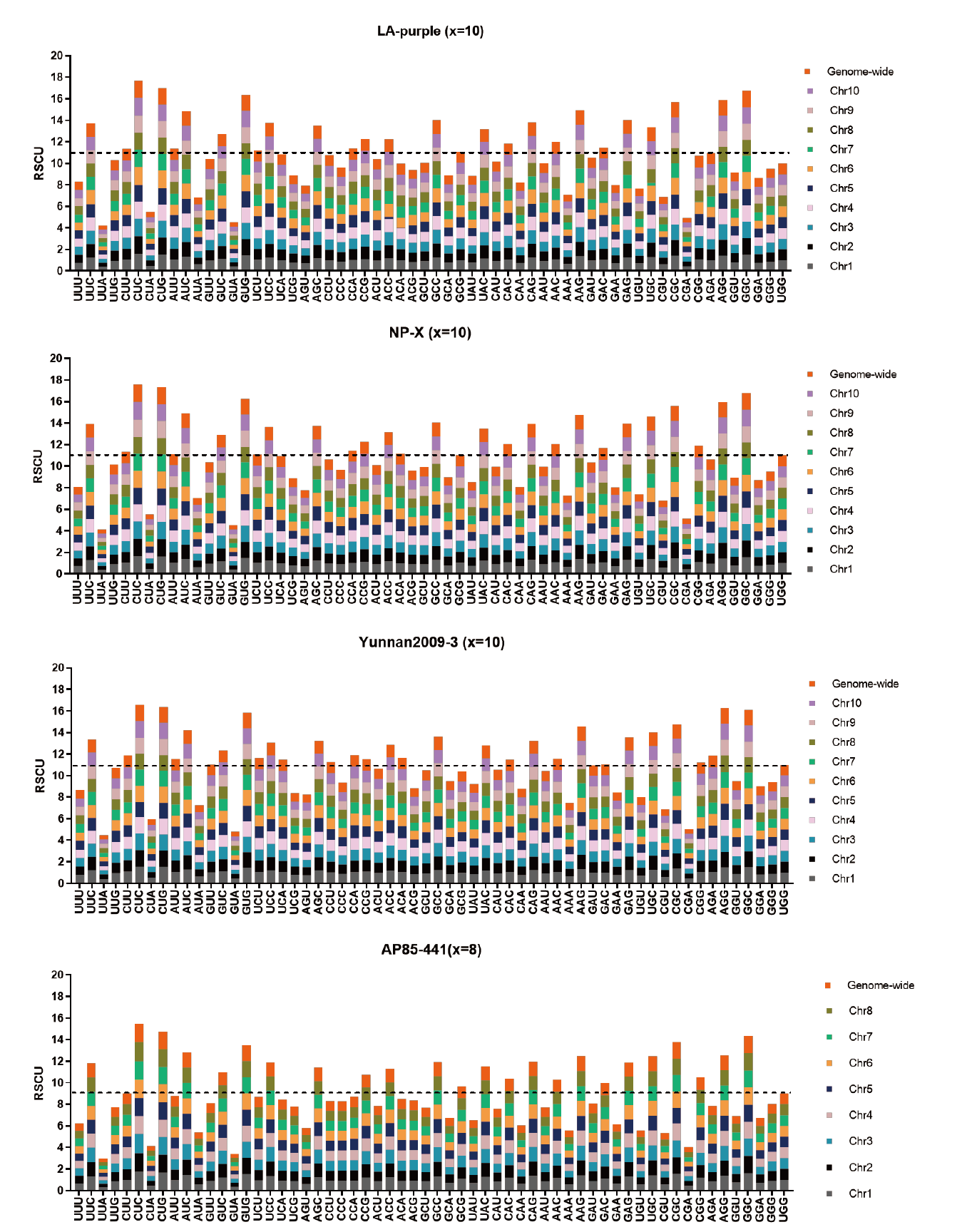
图5 四个材料全基因组和染色体组水平的密码子RSCU值分析 图中虚线表示在全基因组和每个染色体组上密码子的RSCU都为1时的RSCU值。图中RSCU值在虚线以上的密码子即可视为该氨基酸中偏好使用的同义密码子
Fig. 5 Analysis of codon RSCU on genome-wide and chromosome set level in four materials The dotted lines in the figure refers to the RSCU value when RSCU is 1 on both genome-wide level and every chromosome sets. Codons with RSCU above the dotted line in the figures can be regarded as preferred synonymous codons for this amino acid

图7 基于RSCU值的甘蔗与其他7种生物的密码子使用偏好性比较 Ss:甘蔗(甘蔗属种及其近缘属种蔗茅);Zm:玉米;Sb:高粱;Os:水稻;At:拟南芥;Nt:烟草;Ec:大肠杆菌;Sc:酿酒酵母
Fig. 7 Comparison analysis of codon usage bias between sugarcane and other seven organisms based on RSCU value Ss: Saccharum spp.(including Saccharum species and E. fulvus); Zm: Z. mays; Sb: S. bicolor; Os: O. ativa; At: A. thaliana; Nt: N. tabacum; Ec: E. coli; Sc: S. cerevisiae
| [1] |
Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms[J]. Mol Biol Evol, 1985, 2(1): 13-34.
doi: 10.1093/oxfordjournals.molbev.a040335 pmid: 3916708 |
| [2] |
Bulmer M. The selection-mutation-drift theory of synonymous codon usage[J]. Genetics, 1991, 129(3): 897-907.
doi: 10.1093/genetics/129.3.897 pmid: 1752426 |
| [3] |
Hershberg R, Petrov DA. Selection on codon bias[J]. Annu Rev Genet, 2008, 42: 287-299.
doi: 10.1146/annurev.genet.42.110807.091442 pmid: 18983258 |
| [4] | 张乐, 金龙国, 罗玲, 等. 大豆基因组和转录组的核基因密码子使用偏好性分析[J]. 作物学报, 2011, 37(6): 965-974. |
| Zhang L, Jin LG, Luo L, et al. Analysis of nuclear gene codon bias on soybean genome and transcriptome[J]. Acta Agron Sin, 2011, 37(6): 965-974. | |
| [5] | 李亚麒, 黄家雄, 娄予强, 等. 小粒咖啡铁皮卡叶绿体基因组密码子偏好性分析[J]. 西北林学院学报, 2023, 38(2): 92-99. |
| Li YQ, Huang JX, Lou YQ, et al. Codon usage bias of the chloroplast genome in Coffea arabica‘Typica’[J]. J Northwest For Univ, 2023, 38(2): 92-99. | |
| [6] |
Tao P, Dai L, Luo MC, et al. Analysis of synonymous codon usage in classical swine fever virus[J]. Virus Genes, 2009, 38(1): 104-112.
doi: 10.1007/s11262-008-0296-z pmid: 18958611 |
| [7] |
Das S, Paul S, Dutta C. Synonymous codon usage in adenoviruses: influence of mutation, selection and protein hydropathy[J]. Virus Res, 2006, 117(2): 227-236.
pmid: 16307819 |
| [8] |
Yadav MK, Swati D. Comparative genome analysis of six malarial parasites using codon usage bias based tools[J]. Bioinformation, 2012, 8(24): 1230-1239.
doi: 10.6026/97320630081230 pmid: 23275725 |
| [9] | 赖瑞联, 冯新, 陈瑾, 等. 橄榄查尔酮异构酶基因CHI的密码子偏好模式[J]. 应用与环境生物学报, 2017, 23(5): 945-951. |
| Lai RL, Feng X, Chen J, et al. Codon usage pattern of chalcone isomerase gene(CHI)in Canarium album(Lour.) Raeusch[J]. Chin J Appl Environ Biol, 2017, 23(5): 945-951. | |
| [10] |
寇莹莹, 宋英今, 杨少辉, 等. 植酸酶phyA基因的密码子优化及其在大豆中的表达[J]. 作物学报, 2016, 42(12): 1798-1804.
doi: 10.3724/SP.J.1006.2016.01798 |
|
Kou YY, Song YJ, Yang SH, et al. Codon optimization and expression of phyA gene in soybean(Glycine max merr.)[J]. Acta Agron Sin, 2016, 42(12): 1798-1804.
doi: 10.3724/SP.J.1006.2016.01798 URL |
|
| [11] | 周宗梁, 林智敏, 耿丽丽, 等. 水稻中cry1Ah1基因密码子优化方案的比较[J]. 生物工程学报, 2012, 28(10): 1184-1194. |
| Zhou ZL, Lin ZM, Geng LL, et al. Comparison of codon optimizations of cry1Ah1 gene in rice[J]. Chin J Biotechnol, 2012, 28(10): 1184-1194. | |
| [12] | 陈惠, 赵海霞, 王红宁, 等. 植酸酶基因中稀有密码子的改造提高其在毕赤酵母中的表达量[J]. 中国生物化学与分子生物学报, 2005, 21(2): 170-174. |
| Chen H, Zhao HX, Wang HN, et al. Increasing expression level of phytase gene(phyA)in Pichia pastoris by changing rare codons[J]. Chin J Biochem Mol Biol, 2005, 21(2): 170-174. | |
| [13] |
Menzella HG. Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli[J]. Microb Cell Fact, 2011, 10: 15.
doi: 10.1186/1475-2859-10-15 pmid: 21371320 |
| [14] |
Jabeen R, Khan MS, Zafar Y, et al. Codon optimization of cry1Ab gene for hyper expression in plant organelles[J]. Mol Biol Rep, 2010, 37(2): 1011-1017.
doi: 10.1007/s11033-009-9802-1 pmid: 19757171 |
| [15] |
Hirai H, Kashima Y, Hayashi K, et al. Efficient expression of laccase gene from white-rot fungus Schizophyllum commune in a transgenic tobacco plant[J]. FEMS Microbiol Lett, 2008, 286(1): 130-135.
doi: 10.1111/fml.2008.286.issue-1 URL |
| [16] |
D'Hont A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana[J]. Cytogenet Genome Res, 2005, 109(1-3): 27-33.
doi: 10.1159/000082378 pmid: 15753555 |
| [17] |
Zhang JS, Zhang XT, Tang HB, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L[J]. Nat Genet, 2018, 50(11): 1565-1573.
doi: 10.1038/s41588-018-0237-2 |
| [18] |
Zhang Q, Qi YY, Pan HR, et al. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum[J]. Nat Genet, 2022, 54(6): 885-896.
doi: 10.1038/s41588-022-01084-1 pmid: 35654976 |
| [19] |
Wang TY, Wang BY, Hua XT, et al. A complete gap-free diploid genome in Saccharum complex and the genomic footprints of evolution in the highly polyploid Saccharum genus[J]. Nat Plants, 2023, 9(4): 554-571.
doi: 10.1038/s41477-023-01378-0 |
| [20] |
Zhang WJ, Zhou J, Li ZF, et al. Comparative analysis of codon usage patterns among mitochondrion, chloroplast and nuclear genes in Triticum aestivum L[J]. J Integr Plant Biol, 2007, 49(2): 246-254.
doi: 10.1111/jipb.2007.49.issue-2 URL |
| [21] | 冯展, 江媛, 郑燕, 等. 肉苁蓉属植物叶绿体基因组密码子偏好性分析[J]. 中草药, 2023, 54(5): 1540-1550. |
| Feng Z, Jiang Y, Zheng Y, et al. Codon use bias analysis of chloroplast genome of Cistanche[J]. Chin Tradit Herb Drugs, 2023, 54(5): 1540-1550. | |
| [22] |
Wright F. The ‘effective number of codons’ used in a gene[J]. Gene, 1990, 87(1): 23-29.
doi: 10.1016/0378-1119(90)90491-9 pmid: 2110097 |
| [23] |
Jiang Y, Deng F, Wang HL, et al. An extensive analysis on the global codon usage pattern of baculoviruses[J]. Arch Virol, 2008, 153(12): 2273-2282.
doi: 10.1007/s00705-008-0260-1 pmid: 19030954 |
| [24] |
张以忠, 曾文艺, 邓琳琼, 等. 甘蓝S-位点基因SRK、SLG和SP11/SCR密码子偏好性分析[J]. 作物学报, 2022, 48(5): 1152-1168.
doi: 10.3724/SP.J.1006.2022.14003 |
|
Zhang YZ, Zeng WY, Deng LQ, et al. Codon usage bias analysis of S-locus genes SRK, SLG, and SP11/SCR in Brassica oleracea[J]. Acta Agron Sin, 2022, 48(5): 1152-1168.
doi: 10.3724/SP.J.1006.2022.14003 URL |
|
| [25] | 金刚, 覃旭, 龙凌云, 等. 剑麻叶绿体基因组编码序列密码子的使用特征[J]. 福建农林大学学报: 自然科学版, 2018, 47(6): 705-710. |
| Jin G, Qin X, Long LY, et al. Characteristics of codon usage in the chloroplast protein-coding genes of Agave hybrid No.11648[J]. J Fujian Agric For Univ Nat Sci Ed, 2018, 47(6): 705-710. | |
| [26] | 雷佳欣, 张丽娟, 高丹丹, 等. 文冠果基因组密码子偏好性分析[J]. 西北林学院学报, 2023, 38(4): 104-110. |
| Lei JX, Zhang LJ, Gao DD, et al. Codon usage bias of Xanthoceras sorbifolium genome[J]. J Northwest For Univ, 2023, 38(4): 104-110. | |
| [27] |
赖瑞联, 林玉玲, 钟春水, 等. 龙眼生长素受体基因TIR1密码子偏好性分析[J]. 园艺学报, 2016, 43(4): 771-780.
doi: 10.16420/j.issn.0513-353x.2015-0662 |
| Lai RL, Lin YL, Zhong CS, et al. Analysis of codon bias of auxin receptor gene TIR1 in Dimocarpus longan[J]. Acta Hortic Sin, 2016, 43(4): 771-780. | |
| [28] |
Liu QP, Feng Y, Zhao XA, et al. Synonymous codon usage bias in Oryza sativa[J]. Plant Sci, 2004, 167(1): 101-105.
doi: 10.1016/j.plantsci.2004.03.003 URL |
| [29] | 刘汉梅, 何瑞, 张怀渝, 等. 玉米同义密码子偏爱性分析[J]. 农业生物技术学报, 2010, 18(3): 456-461. |
| Liu HM, He R, Zhang HY, et al. Analysis of synonymous codon bias in maize[J]. J Agric Biotechnol, 2010, 18(3): 456-461. | |
| [30] | 张晓峰, 薛庆中. 水稻和拟南芥NBS-LRR基因家族同义密码子使用偏好的比较[J]. 作物学报, 2005, 31(5): 596-602. |
| Zhang XF, Xue QZ. Synonymous codon bias of NBS-LRR gene family in rice and Arabidopsis[J]. Acta Agron Sin, 2005, 31(5): 596-602. | |
| [31] |
Kawabe A, Miyashita NT. Patterns of codon usage bias in three dicot and four monocot plant species[J]. Genes Genet Syst, 2003, 78(5): 343-352.
doi: 10.1266/ggs.78.343 pmid: 14676425 |
| [32] |
Comeron JM, Aguadé M. An evaluation of measures of synonymous codon usage bias[J]. J Mol Evol, 1998, 47(3): 268-274.
doi: 10.1007/pl00006384 pmid: 9732453 |
| [33] |
唐玉娟, 赵英, 黄国弟, 等. 芒果叶绿体基因组密码子使用偏好性分析[J]. 热带作物学报, 2021, 42(8): 2143-2150.
doi: 10.3969/j.issn.1000-2561.2021.08.004 |
| Tang YJ, Zhao Y, Huang GD, et al. Analysis on codon usage bias of chloroplast genes from mango[J]. Chin J Trop Crops, 2021, 42(8): 2143-2150. | |
| [34] |
杨祥燕, 蔡元保, 谭秦亮, 等. 菠萝叶绿体基因组密码子偏好性分析[J]. 热带作物学报, 2022, 43(3): 439-446.
doi: 10.3969/j.issn.1000-2561.2022.03.001 |
| Yang XY, Cai YB, Tan QL, et al. Analysis of codon usage bias in the chloroplast genome of Ananas comosus[J]. Chin J Trop Crops, 2022, 43(3): 439-446. | |
| [35] |
Wen Y, Zou ZL, Li HS, et al. Analysis of codon usage patterns in Morus notabilis based on genome and transcriptome data[J]. Genome, 2017, 60(6): 473-484.
doi: 10.1139/gen-2016-0129 URL |
| [36] | 王占军, 丁亮, 蔡倩文, 等. 3种木薯全基因组的密码子偏好性模式与变异来源比较[J]. 应用与环境生物学报, 2021, 27(4): 1013-1021. |
| Wang ZJ, Ding L, Cai QW, et al. Comparison of codon preference patterns and variation sources in Manihot esculenta Crantz genomes[J]. Chin J Appl Environ Biol, 2021, 27(4): 1013-1021. | |
| [37] | 毛立彦, 黄秋伟, 龙凌云, 等. 7种睡莲属植物叶绿体基因组密码子偏好性分析[J]. 西北林学院学报, 2022, 37(2): 98-107. |
| Mao LY, Huang QW, Long LY, et al. Comparative analysis of codon usage bias in chloroplast genomes of seven Nymphaea species[J]. J Northwest For Univ, 2022, 37(2): 98-107. | |
| [38] | 冯瑞云, 梅超, 王慧杰, 等. 籽粒苋叶绿体基因组密码子偏好性分析[J]. 中国草地学报, 2019, 41(4): 8-15. |
| Feng RY, Mei C, Wang HJ, et al. Analysis of codon usage in the chloroplast genome of grain amaranth(Amaranthus hypochondriacus L.)[J]. Chin J Grassland, 2019, 41(4): 8-15. | |
| [39] | 原晓龙, 李云琴, 张劲峰, 等. 乳油木叶绿体基因组密码子偏好性分析[J]. 分子植物育种, 2020, 18(17): 5658-5664. |
| Yuan XL, Li YQ, Zhang JF, et al. Codon usage bias analysis of chloroplast genome in Vitellaria paradoxa[J]. Mol Plant Breed, 2020, 18(17): 5658-5664. | |
| [40] |
Tang DF, Wei F, Cai ZQ, et al. Analysis of codon usage bias and evolution in the chloroplast genome of Mesona chinensis Benth[J]. Dev Genes Evol, 2021, 231(1-2): 1-9.
doi: 10.1007/s00427-020-00670-9 |
| [1] | 王欣, 徐一亿, 徐扬, 徐辰武. 作物全基因组选择育种技术研究进展[J]. 生物技术通报, 2024, 40(3): 1-13. |
| [2] | 杨伟成, 孙岩, 杨倩, 王壮琳, 马菊花, 薛金爱, 李润植. 陆地棉FAX家族的全基因组鉴定及GhFAX1的功能分析[J]. 生物技术通报, 2024, 40(3): 155-169. |
| [3] | 龚丽丽, 余花, 杨杰, 陈天池, 赵双滢, 吴月燕. 葡萄CYP707A基因家族的鉴定及对果实成熟的功能验证[J]. 生物技术通报, 2024, 40(2): 160-171. |
| [4] | 路喻丹, 刘晓驰, 冯新, 陈桂信, 陈义挺. 猕猴桃BBX基因家族成员鉴定与转录特征分析[J]. 生物技术通报, 2024, 40(2): 172-182. |
| [5] | 杨雨青, 谭娟, 汪芳, 彭顺利, 陈婕, 谭明燕, 吕美艳, 周富裕, 刘声传. 茶树叶绿体基因组的研究与应用进展[J]. 生物技术通报, 2024, 40(2): 20-30. |
| [6] | 周会汶, 吴兰花, 韩德鹏, 郑伟, 余跑兰, 吴杨, 肖小军. 甘蓝型油菜种子硫苷含量全基因组关联分析[J]. 生物技术通报, 2024, 40(1): 222-230. |
| [7] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [8] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [9] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [10] | 饶紫环, 谢志雄. 一株Olivibacter jilunii 纤维素降解菌株的分离鉴定与降解能力分析[J]. 生物技术通报, 2023, 39(8): 283-290. |
| [11] | 郭少华, 毛会丽, 刘征权, 付美媛, 赵平原, 马文博, 李旭东, 关建义. 一株鱼源致病性嗜水气单胞菌XDMG的全基因组测序及比较基因组分析[J]. 生物技术通报, 2023, 39(8): 291-306. |
| [12] | 杜冬冬, 钱晶, 李思琪, 刘雯菲, 魏向利, 刘长勇, 罗瑞峰, 康立超. 单核细胞增生李斯特菌LMXJ15全基因组测序及分析[J]. 生物技术通报, 2023, 39(7): 298-306. |
| [13] | 李雨真, 梅天秀, 李治文, 王淇, 李俊, 邹岳, 赵心清. 红酵母基因组和代谢工程改造研究进展[J]. 生物技术通报, 2023, 39(7): 67-79. |
| [14] | 尹明华, 余锾媛, 肖心怡, 王玉婷. 江西铅山红芽芋叶绿体基因组特征及系统发育分析[J]. 生物技术通报, 2023, 39(6): 233-247. |
| [15] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||