








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 82-91.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0460
• 薯类作物生物技术专题(专题主编:徐建飞,尚轶) • 上一篇 下一篇
吴娟1( ), 武小娟1, 王沛捷1, 谢锐2, 聂虎帅1, 李楠1, 马艳红1(
), 武小娟1, 王沛捷1, 谢锐2, 聂虎帅1, 李楠1, 马艳红1( )
)
收稿日期:2024-05-17
出版日期:2024-09-26
发布日期:2024-10-12
通讯作者:
马艳红,女,博士,教授,研究方向:马铃薯品质性状调控机制;E-mail: mayanhong80@126.com作者简介:吴娟,女,博士研究生,研究方向:马铃薯品质性状调控机制;E-mail: 1977389659@qq.com
基金资助:
WU Juan1( ), WU Xiao-juan1, WANG Pei-jie1, XIE Rui2, NIE Hu-shuai1, LI Nan1, MA Yan-hong1(
), WU Xiao-juan1, WANG Pei-jie1, XIE Rui2, NIE Hu-shuai1, LI Nan1, MA Yan-hong1( )
)
Received:2024-05-17
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】乙烯响应因子(ethylene responsive factor,ERF)是一类重要的转录因子,参与调控植物花青素的生物合成。筛选可能参与调控马铃薯块茎花青素合成的StERFs基因,为开展彩色马铃薯花青素相关StERFs基因功能研究奠定基础。【方法】利用生物信息学方法对其理化性质、亚细胞定位、保守基序、进化关系、蛋白质二级结构、启动子顺式作用元件及蛋白互作关系进行分析,同时采用RT-qPCR方法对不同颜色马铃薯薯肉中StERFs进行表达模式分析。【结果】基于不同颜色马铃薯块茎的转录组表达谱,获得7个差异表达ERF转录因子。7个StERFs属于亲水性蛋白,均预测定位于细胞核。Motif 1(RWLG)和Motif 2(YRG)是7个StERFs中共同的保守基序,属于AP2的结构域特征序列。StERFs基因启动子序列含有响应激素作用元件,以及响应防御与胁迫、低温和光照等的顺式作用元件。7个StERFs在紫色薯肉中表达量显著高于黄色薯肉,其中StERF72和StERF110与已知的花青素相关ERFs(IbERF71和PyERF3)亲缘关系较近,且7个StERFs与56个转录因子存在蛋白互作关系。【结论】7个StERFs基因可能参与调控马铃薯块茎中花青素的合成,其中StERF72和StERF110可作为候选基因进一步验证其功能。
吴娟, 武小娟, 王沛捷, 谢锐, 聂虎帅, 李楠, 马艳红. 彩色马铃薯花青素合成相关ERF基因筛选及表达分析[J]. 生物技术通报, 2024, 40(9): 82-91.
WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato[J]. Biotechnology Bulletin, 2024, 40(9): 82-91.
| 基因序号Gene ID | 基因名称Gene name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| Soltu.DM.05G020900 | StERF92-1 | F-ATGGAGTTAGGGTTTGGCTAGGAAC R-CCCTCATTGATAATGCGGCTTGATC |
| Soltu.DM.09G021200 | StERF92-2 | F-CTGAAGAAGCTGCGTTAGCGTATG R-ACCGGCGAACAATTCTCATCAAATC |
| Soltu.DM.09G023730 | StERF72 | F-GAAAAGGGGTTAGGGTCTGGTTAGG R-AGTAGTGGTCGTCGTCTTCATTGG |
| Soltu.DM.04G021630 | StERF059 | F-TGCATCAACAACAACAGAGCTTAGG R-GTTTGCTTCATCGGGACTGGTTTC |
| Soltu.DM.04G027450 | StEF110 | F-TGATGACGCCGCTCTTAGATTCAG R-TGGTGCTGGTGCTGTGGTTG |
| Soltu.DM.03G031310 | StERF3 | F-TGCCACATGACATCTCTCACTGC R-TCTCACTCTGCCTTTCTTCTCCATC |
| Soltu.DM.04G033390 | StERF10 | F-CAGAGGAATACGGATGAGGAAGTGG R-GTAAGCACGAGCAGCAGCAAC |
| Actin | F-CCTTGTATGCTAGTGGTCG R-GCTCATAGTCAAGAGCCAC |
表1 RT-qPCR引物序列
Table 1 Primer sequences for RT-qPCR
| 基因序号Gene ID | 基因名称Gene name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| Soltu.DM.05G020900 | StERF92-1 | F-ATGGAGTTAGGGTTTGGCTAGGAAC R-CCCTCATTGATAATGCGGCTTGATC |
| Soltu.DM.09G021200 | StERF92-2 | F-CTGAAGAAGCTGCGTTAGCGTATG R-ACCGGCGAACAATTCTCATCAAATC |
| Soltu.DM.09G023730 | StERF72 | F-GAAAAGGGGTTAGGGTCTGGTTAGG R-AGTAGTGGTCGTCGTCTTCATTGG |
| Soltu.DM.04G021630 | StERF059 | F-TGCATCAACAACAACAGAGCTTAGG R-GTTTGCTTCATCGGGACTGGTTTC |
| Soltu.DM.04G027450 | StEF110 | F-TGATGACGCCGCTCTTAGATTCAG R-TGGTGCTGGTGCTGTGGTTG |
| Soltu.DM.03G031310 | StERF3 | F-TGCCACATGACATCTCTCACTGC R-TCTCACTCTGCCTTTCTTCTCCATC |
| Soltu.DM.04G033390 | StERF10 | F-CAGAGGAATACGGATGAGGAAGTGG R-GTAAGCACGAGCAGCAGCAAC |
| Actin | F-CCTTGTATGCTAGTGGTCG R-GCTCATAGTCAAGAGCCAC |
| 基因序号 Gene ID | 基因名称 Gene name | NCBI数据库BLAST序列ID NCBI database BLAST sequence ID | PlantTB数据库BLAST序列ID PlantTB database BLAST sequence ID | 相似度 Identities/% |
|---|---|---|---|---|
| Soltu.DM.05G020900 | StERF92-1 | XP_006360378.1 | PGSC0003DMP400025705 | 100 |
| Soltu.DM.09G021200 | StERF92-2 | XP_006351652.2 | PGSC0003DMP400018171 | 100 |
| Soltu.DM.09G023730 | StERF72 | NP_001275232.1 | PGSC0003DMP400055026 | 92 |
| Soltu.DM.04G021630 | StERF059 | XP_006355363.1 | PGSC0003DMP400035842 | 100 |
| Soltu.DM.04G027450 | StEF110 | XP_006363442.1 | PGSC0003DMP400011298 | 100 |
| Soltu.DM.03G031310 | StERF3 | XP_006341371.1 | PGSC0003DMP400024835 | 92 |
| Soltu.DM.04G033390 | StERF10 | XP_006354856.1 | PGSC0003DMP400013982 | 100 |
表2 同源序列比对
Table 2 An alignment of the homologous sequences
| 基因序号 Gene ID | 基因名称 Gene name | NCBI数据库BLAST序列ID NCBI database BLAST sequence ID | PlantTB数据库BLAST序列ID PlantTB database BLAST sequence ID | 相似度 Identities/% |
|---|---|---|---|---|
| Soltu.DM.05G020900 | StERF92-1 | XP_006360378.1 | PGSC0003DMP400025705 | 100 |
| Soltu.DM.09G021200 | StERF92-2 | XP_006351652.2 | PGSC0003DMP400018171 | 100 |
| Soltu.DM.09G023730 | StERF72 | NP_001275232.1 | PGSC0003DMP400055026 | 92 |
| Soltu.DM.04G021630 | StERF059 | XP_006355363.1 | PGSC0003DMP400035842 | 100 |
| Soltu.DM.04G027450 | StEF110 | XP_006363442.1 | PGSC0003DMP400011298 | 100 |
| Soltu.DM.03G031310 | StERF3 | XP_006341371.1 | PGSC0003DMP400024835 | 92 |
| Soltu.DM.04G033390 | StERF10 | XP_006354856.1 | PGSC0003DMP400013982 | 100 |
| 蛋白名称 Protein name | 氨基酸数目 Number of amion acids/aa | 相对分子质量 Molecular weight/ kD | 理论等电点 pI | 分子式 Molecular formula | 脂肪系数 Aliphatc index | 总平均亲水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| StERF92-1 | 225 | 25 615.43 | 5.4 | C1125H1749N311O364S5 | 70.18 | -0.738 | 细胞核 |
| StERF92-2 | 197 | 22 839.51 | 4.89 | C1003H1554N276O319S8 | 68.78 | -0.652 | 细胞核 |
| StERF72 | 278 | 31 714.03 | 5.1 | C1385H2129N391O448S9 | 57.55 | -0.905 | 细胞核 |
| StERF059 | 370 | 40 579.22 | 5.61 | C1785H2762N480O576S13 | 66.84 | -0.465 | 细胞核 |
| StEF110 | 441 | 48 254.51 | 5.97 | C2084H3186N612O695S10 | 46.78 | -0.834 | 细胞核 |
| StERF3 | 159 | 18 282.48 | 6.91 | C782H1250N234O254S9 | 59.62 | -0.899 | 细胞核 |
| StERF10 | 145 | 16 663.74 | 9.04 | C722H1139N219O222S7 | 56.62 | -0.974 | 细胞核 |
表3 马铃薯StERFs理化性质分析及亚细胞定位预测
Table 3 Analysis of physicochemical properties and prediction of subcellular localization in potato StERFs
| 蛋白名称 Protein name | 氨基酸数目 Number of amion acids/aa | 相对分子质量 Molecular weight/ kD | 理论等电点 pI | 分子式 Molecular formula | 脂肪系数 Aliphatc index | 总平均亲水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| StERF92-1 | 225 | 25 615.43 | 5.4 | C1125H1749N311O364S5 | 70.18 | -0.738 | 细胞核 |
| StERF92-2 | 197 | 22 839.51 | 4.89 | C1003H1554N276O319S8 | 68.78 | -0.652 | 细胞核 |
| StERF72 | 278 | 31 714.03 | 5.1 | C1385H2129N391O448S9 | 57.55 | -0.905 | 细胞核 |
| StERF059 | 370 | 40 579.22 | 5.61 | C1785H2762N480O576S13 | 66.84 | -0.465 | 细胞核 |
| StEF110 | 441 | 48 254.51 | 5.97 | C2084H3186N612O695S10 | 46.78 | -0.834 | 细胞核 |
| StERF3 | 159 | 18 282.48 | 6.91 | C782H1250N234O254S9 | 59.62 | -0.899 | 细胞核 |
| StERF10 | 145 | 16 663.74 | 9.04 | C722H1139N219O222S7 | 56.62 | -0.974 | 细胞核 |
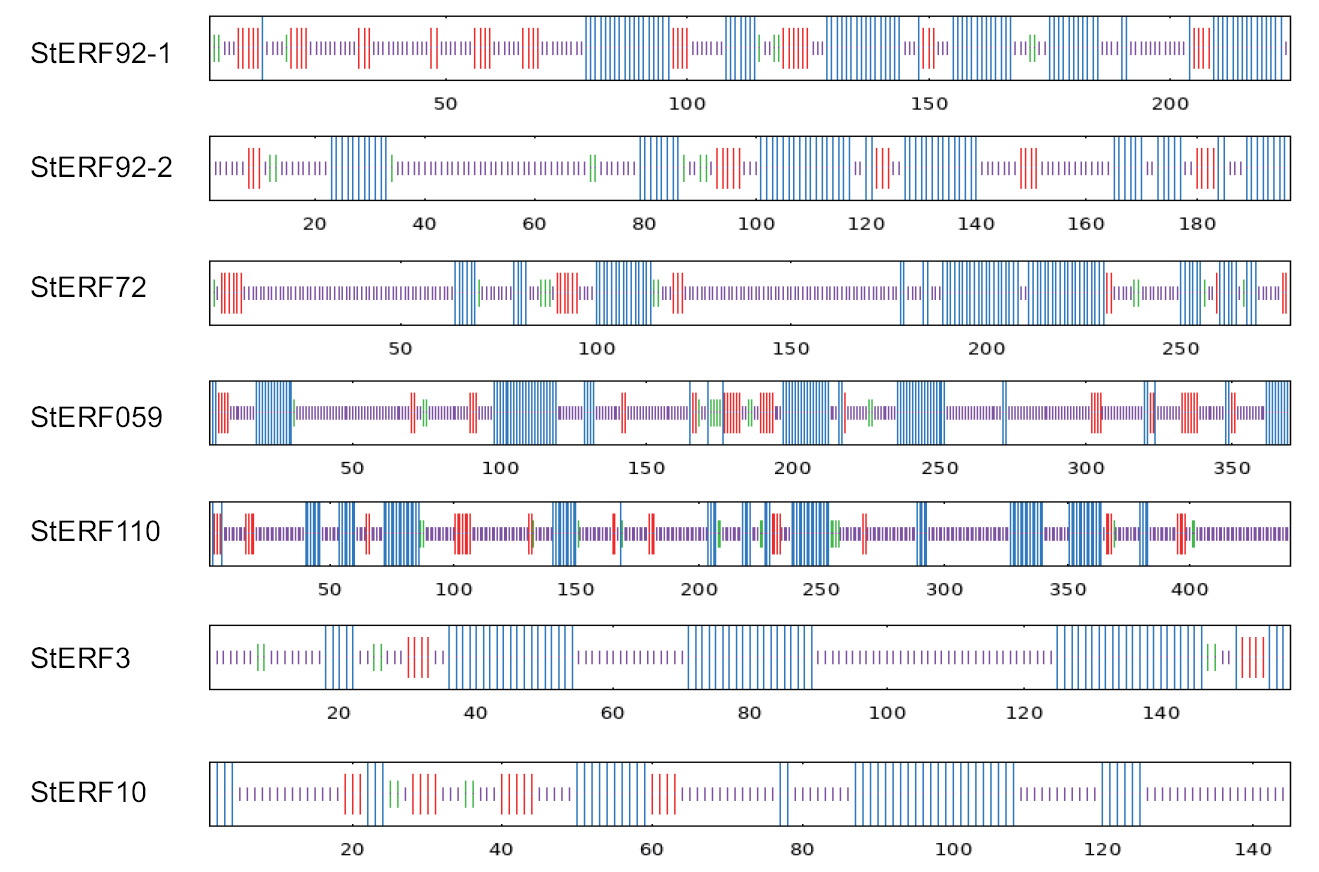
图4 马铃薯StERFs蛋白二级结构 图中蓝色为α-螺旋;紫色为无规则卷曲;红色为延伸链;绿色为β-折叠
Fig. 4 Secondary structure of the potato StERFs protein The figure shows α-helices in blue; irregular coiling in purple; extended chains in red; and β-folding in green

图5 马铃薯StERFs及其他已知的花青素相关ERFs的氨基酸序列比对 图中红色的方框为AP2结构域中典型的YRG和WLG单元
Fig. 5 Amino acid sequence alignment of potato StERFs and other known anthocyanin-related ERFs The red boxes in the figure show typical YRG and WLG units in the AP2 structural domains
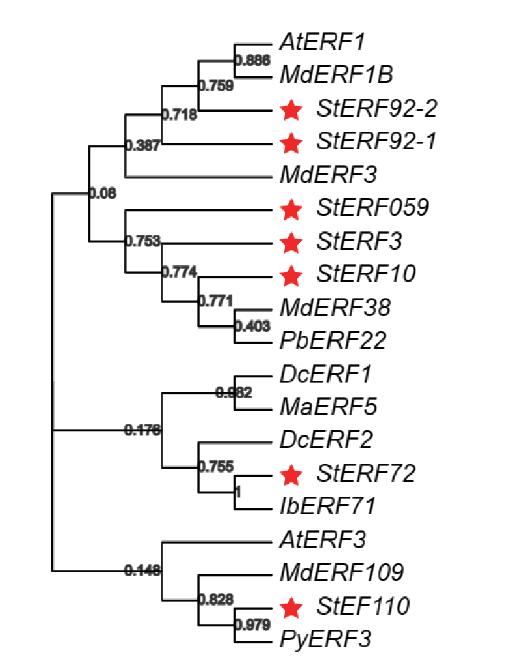
图6 马铃薯StERFs及其他已知的花青素相关ERFs的系统进化树 图中红色五角星为转录组筛选到的7个马铃薯StERFs
Fig. 6 Phylogenetic tree of potato StERFs and other known anthocyanin-related ERFs The red pentagons in the figure show the seven potato StERFs screened by the transcriptome
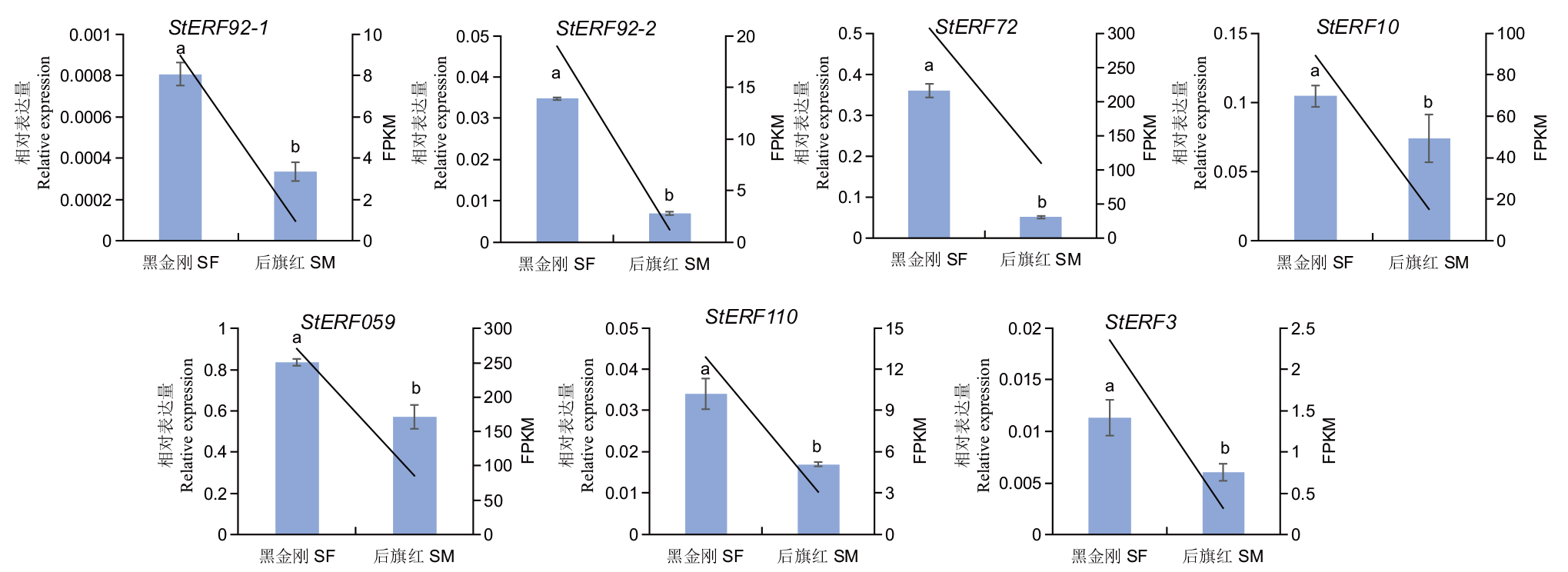
图9 彩色马铃薯StERFs表达模式 柱形代表通过RT-qPCR测定的基因相对表达量,折线代表通过转录组测序的基因FPKM值。不同小写字母表示P<0.05
Fig. 9 Expression pattern of StERFs in colored potato Bars indicate gene relative expression as determined by RT-qPCR, and broken lines indicate gene FPKM values by transcriptome sequencing. Different lower letters indicate significant difference at 0.05 level
| [1] | He J, Giusti MM. Anthocyanins: natural colorants with health-promoting properties[J]. Annu Rev Food Sci Technol, 2010, 1: 163-187. |
| [2] |
Echeverry C, Arredondo F, Martínez M, et al. Antioxidant activity, cellular bioavailability, and iron and calcium management of neuroprotective and nonneuroprotective flavones[J]. Neurotox Res, 2015, 27(1): 31-42.
doi: 10.1007/s12640-014-9483-y pmid: 24972590 |
| [3] |
Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology[J]. Plant Physiol, 2001, 126(2): 485-493.
doi: 10.1104/pp.126.2.485 pmid: 11402179 |
| [4] |
Zoratti L, Karppinen K, Luengo Escobar A, et al. Light-controlled flavonoid biosynthesis in fruits[J]. Front Plant Sci, 2014, 5: 534.
doi: 10.3389/fpls.2014.00534 pmid: 25346743 |
| [5] | Licausi F, Giorgi FM, Zenoni S, et al. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera[J]. BMC Genomics, 2010, 11: 719. |
| [6] |
Sharoni AM, Nuruzzaman M, Satoh K, et al. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice[J]. Plant Cell Physiol, 2011, 52(2): 344-360.
doi: 10.1093/pcp/pcq196 pmid: 21169347 |
| [7] | An JP, Zhang XW, Bi SQ, et al. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple[J]. Plant J, 2020, 101(3): 573-589. |
| [8] |
Zhang J, Xu HF, Wang N, et al. The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple[J]. Plant Mol Biol, 2018, 98(3): 205-218.
doi: 10.1007/s11103-018-0770-5 pmid: 30182194 |
| [9] | An JP, Wang XF, Li YY, et al. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation[J]. Plant Physiol, 2018, 178(2): 808-823. |
| [10] | Ma HY, Yang T, Li Y, et al. The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit[J]. Plant Cell, 2021, 33(10): 3309-3330. |
| [11] | Mo RL, Han GM, Zhu ZX, et al. The ethylene response factor ERF5 regulates anthocyanin biosynthesis in ‘zijin’ mulberry fruits by interacting with MYBA and F3H genes[J]. Int J Mol Sci, 2022, 23(14): 7615. |
| [12] | Cheng MC, Liao PM, Kuo WW, et al. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals[J]. Plant Physiol, 2013, 162(3): 1566-1582. |
| [13] | Ning ZY, Hu KD, Zhou ZL, et al. IbERF71, with IbMYB340 and IbbHLH2, coregulates anthocyanin accumulation by binding to the IbANS1 promoter in purple-fleshed sweet potato(Ipomoea batatas L.)[J]. Plant Cell Rep, 2021, 40(1): 157-169. |
| [14] | Yao GF, Ming ML, Allan AC, et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis[J]. Plant J, 2017, 92(3): 437-451. |
| [15] | Wu T, Liu HT, Zhao GP, et al. Jasmonate and ethylene-regulated ethylene response factor 22 promotes lanolin-induced anthocyanin biosynthesis in ‘zaosu’ pear(Pyrusbretschneideri rehd.) fruit[J]. Biomolecules, 2020, 10(2): 278. |
| [16] | Kimura S, Chikagawa Y, Kato M, et al. Upregulation of the promoter activity of the carrot(Daucus carota)phenylalanine ammonia-lyase gene(DcPAL3)is caused by new members of the transcriptional regulatory proteins, DcERF1 and DcERF2, which bind to the GCC-box homolog and act as an activator to the DcPAL3 promoter[J]. J Plant Res, 2008, 121(5): 499-508. |
| [17] |
Stokstad E. The new potato[J]. Science, 2019, 363(6427): 574-577.
doi: 10.1126/science.363.6427.574 pmid: 30733400 |
| [18] |
Huang XH, Han B. The magic of genomics in creating hybrid potato[J]. Mol Plant, 2021, 14(8): 1237-1238.
doi: 10.1016/j.molp.2021.06.015 pmid: 34166848 |
| [19] | 白嗣鲜, 刘斌, 吕明举, 等. 内蒙古自治区马铃薯种业现状及发展趋势分析[J]. 现代农业, 2022,(5): 56-58. |
| Bai SX, Liu B, Lyu MJ, et al. Analysis of the current situation and development trend of potato seed industry in Inner Mongolia Autonomous Region[J]. Modern Agriculture, 2022,(5): 56-58. | |
| [20] | 齐恩芳, 贾小霞, 黄伟, 等. 彩色马铃薯花色素苷生物合成及其调控研究进展[J]. 中国蔬菜, 2023,(7): 25-32. |
| Qi EF, Jia XX, Huang W, et al. Progress in the biosynthesis and control of anthocyanin glycosides in colored potatoes[J]. Chinese Vegetable, 2023,(7): 25-32. | |
| [21] | 赵营, 刘维刚, 李世贵, 等. 15个马铃薯AP2/ERF转录因子的生物信息学与表达分析[J]. 分子植物育种, 2023, 21(9): 2806-2815. |
| Zhao Y, Liu WG, Li SG, et al. Bioinformatics and expression analysis of 15 potato AP2/ERF transcription factors[J]. Mol Plant Breed, 2023, 21(9): 2806-2815. | |
| [22] | 王睿, 濮雪, 王凯彤, 等. 马铃薯StERF5基因的生物信息学及表达分析[J]. 农业生物技术学报, 2023, 31(2): 250-258. |
| Wang R, Pu X, Wang KT, et al. Bioinformatics and expression analysis of StERF5 gene in potato(Solanum tuberosum)[J]. J Agric Biotechnol, 2023, 31(2): 250-258. | |
| [23] | 王芳芳, 杨江伟, 朱熙, 等. 马铃薯StERF109基因的生物信息学及表达分析[J]. 农业生物技术学报, 2021, 29(11): 2087-2098. |
| Wang FF, Yang JW, Zhu X, et al. Bioinformatics and expression analysis of StERF109 gene in potato(Solanum tuberosum)[J]. J Agric Biotechnol, 2021, 29(11): 2087-2098. | |
| [24] | Wu J, Ma YH, Xie R, et al. Integrated metabolomics and transcriptomics reveals difference involved in flavonoid and indole alkaloids biosynthesis in potato tuber flesh[J]. Sci Hortic, 2024, 324: 112630. |
| [25] | Varet H, Brillet-Guéguen L, Coppée JY, et al. SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data[J]. PLoS One, 2016, 11(6): e0157022. |
| [26] |
Dheda K, Huggett JF, Bustin SA, et al. Validation of housekeeping genes for normalizing RNA expression in real-time PCR[J]. BioTechniques, 2004, 37(1): 112-114, 116, 118-119.
pmid: 15283208 |
| [27] |
Feng K, Hou XL, Xing GM, et al. Advances in AP2/ERF super-family transcription factors in plant[J]. Crit Rev Biotechnol, 2020, 40(6): 750-776.
doi: 10.1080/07388551.2020.1768509 pmid: 32522044 |
| [28] |
赵雪惠, 王庆杰, 李晨, 等. 桃ERF转录因子家族生物信息学分析及芽萌发相关基因筛选[J]. 植物学报, 2018, 53(5): 612-624.
doi: 10.11983/CBB17117 |
| Zhao XH, Wang QJ, Li C, et al. Genome-wide identification of ethylene responsive factor(ERF)family genes in peach and screening of genes related to germination[J]. Chin Bull Bot, 2018, 53(5): 612-624. | |
| [29] | 卞云迪, 张驰, 王雪晴, 等. 小麦AP2/ERF转录因子家族生物信息学分析[J]. 天津师范大学学报: 自然科学版, 2022, 42(4): 39-45. |
| Bian YD, Zhang C, Wang XQ, et al. Bioinformatics analysis of TaAP2/ERF transcription factor family in wheat[J]. J Tianjin Norm Univ Nat Sci Ed, 2022, 42(4): 39-45. | |
| [30] |
吴婧, 范菠菠, 张学峰, 等. 蒙古冰草ERF转录因子生物信息学及其表达分析[J]. 草地学报, 2022, 30(11): 2910-2921.
doi: 10.11733/j.issn.1007-0435.2022.11.008 |
| Wu J, Fan BB, Zhang XF, et al. Bioinformatics and expression analysis of ERF transcription factor in agropy ron mongolicum Keng[J]. Acta Agrestia Sin, 2022, 30(11): 2910-2921. | |
| [31] | 张计育, 王庆菊, 郭忠仁. 植物AP2/ERF 类转录因子研究进展[J]. 遗传, 2012, 34(7): 835-847. |
| Zhang JY, Wang QJ, Guo ZR. Progresses on plant AP2/ERF transcription factors[J]. Hereditas(Beijing), 2012, 34(7): 835-847. | |
| [32] | Abou El-Dis GR, Zavdetovna KL, Nikolaevich AA, et al. Influence of light on the accumulation of anthocyanins in callus culture of Vaccinium corymbosum L. cv. Sunt Blue Giant[J]. J Photochem Photobiol, 2021, 8: 100058. |
| [33] | Mao WW, Han Y, Chen YT, et al. Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of Chalcone Synthase 1[J]. Plant Cell, 2022, 34(4): 1226-1249. |
| [34] | Shahab M, Roberto SR, Ahmed S, et al. Relationship between anthocyanins and skin color of table grapes treated with abscisic acid at different stages of berry ripening[J]. Sci Hortic, 2020, 259: 108859. |
| [35] | Fang X, Zhang LZ, Wang LJ. The transcription factor MdERF78 is involved in ALA-induced anthocyanin accumulation in apples[J]. Front Plant Sci, 2022, 13: 915197. |
| [36] | Ni JB, Bai SL, Zhao Y, et al. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114[J]. Plant Mol Biol, 2019, 99(1-2): 67-78. |
| [1] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [2] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| [3] | 王青, 倪尔冬, 王秋霜, 秦丹丹, 方开星, 李红建, 姜晓辉, 李波, 潘晨东, 吴华玲. 植物花青素修饰相关UDP-糖基转移酶研究进展[J]. 生物技术通报, 2024, 40(7): 28-42. |
| [4] | 胡锦锦, 李素贞, 马旭辉, 柳小庆, 谢珊珊, 江海洋, 陈茹梅. 玉米花青素生物合成代谢调控[J]. 生物技术通报, 2024, 40(6): 34-44. |
| [5] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [6] | 王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218. |
| [7] | 吴泽航, 杨中义, 鄢毅铖, 贾永红, 吴月燕, 谢晓鸿. 比利时杜鹃花类黄酮3'-羟化酶(F3'H)基因克隆及功能分析[J]. 生物技术通报, 2024, 40(6): 251-259. |
| [8] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [9] | 杜兵帅, 邹昕蕙, 王子豪, 张馨元, 曹一博, 张凌云. 油茶SWEET基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 179-190. |
| [10] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [11] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [12] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [13] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [14] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [15] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||