








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 34-44.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0075
胡锦锦1( ), 李素贞2, 马旭辉2, 柳小庆2, 谢珊珊1, 江海洋1(
), 李素贞2, 马旭辉2, 柳小庆2, 谢珊珊1, 江海洋1( ), 陈茹梅2(
), 陈茹梅2( )
)
收稿日期:2024-01-17
出版日期:2024-06-26
发布日期:2024-06-24
通讯作者:
陈茹梅,女,研究员,研究方向:植物分子生物学与基因工程;E-mail: chenrumei@caas.cn;作者简介:胡锦锦,女,硕士研究生,研究方向:植物分子生物学;E-mail: h248229@163.com
基金资助:
HU Jin-jin1( ), LI Su-zhen2, MA Xu-hui2, LIU Xiao-qing2, XIE Shan-shan1, JIANG Hai-yang1(
), LI Su-zhen2, MA Xu-hui2, LIU Xiao-qing2, XIE Shan-shan1, JIANG Hai-yang1( ), CHEN Ru-mei2(
), CHEN Ru-mei2( )
)
Received:2024-01-17
Published:2024-06-26
Online:2024-06-24
摘要:
花青素(anthocyanidin)以糖苷形式与葡萄糖或纤维二糖分子结合存在,是一类水溶性天然色素,广泛存在于自然界中的果蔬和花卉中。花青素具有抗氧化、抗炎、抗肿瘤以及抑制病毒生长等生理活性,因此在食品和医药领域具有广阔的应用前景。随着对植物花青素合成途径的深入研究,花青素生物合成途径逐渐清晰,但是花青素调控基因以及机制仍有很大的探索空间。紫玉米是一种富含花青素的作物,其花青素生物合成途径的研究具有重要的实际意义。近年来,研究人员在富含花青素玉米研究中取得了较大进展,但在花青素玉米的精准育种方面有待进一步研究。本文从玉米的花青素生物合成途径、关键酶基因和调控基因等方面进行综述,系统分析了玉米花青素合成途径中各个结构基因以及MYB、bHLH、WD40等调控基因对花青素合成的影响;阐述了MBW三元复合物对花青素的调控;讨论了富含花青素玉米育种中尚未解决的问题及未来研究方向,以期为今后开展花青素玉米遗传改良以及选育高花青素玉米品种提供参考。
胡锦锦, 李素贞, 马旭辉, 柳小庆, 谢珊珊, 江海洋, 陈茹梅. 玉米花青素生物合成代谢调控[J]. 生物技术通报, 2024, 40(6): 34-44.
HU Jin-jin, LI Su-zhen, MA Xu-hui, LIU Xiao-qing, XIE Shan-shan, JIANG Hai-yang, CHEN Ru-mei. Regulation of Maize Anthocyanin Biosynthesis Metabolism[J]. Biotechnology Bulletin, 2024, 40(6): 34-44.
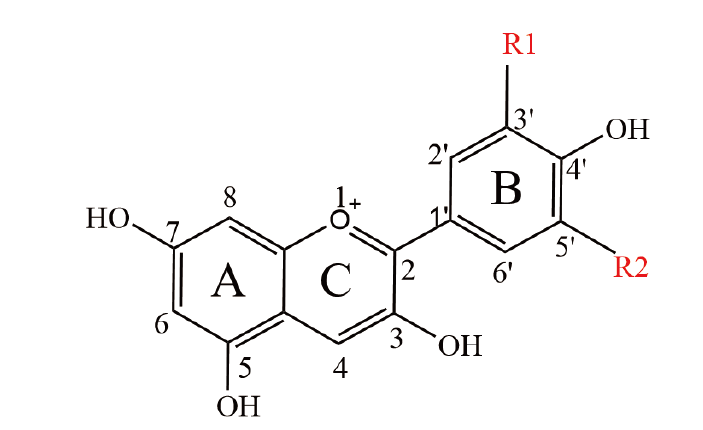
图1 花青素的化学结构 A-C指代苯环,1-8为A、C环碳位号,1'-6'为B环碳位号
Fig. 1 Chemical structure of anthocyanins A-C refer to the benzene ring, 1-8 are A and C ring carbon numbers, 1'-6' are B ring carbon numbers

图2 花青素生物合成途径 PAL:苯丙氨酸裂解酶; C4H:肉桂酸羟化酶; 4CL:香豆酸 CoA 连接酶;CHS:查尔酮合成酶;CHI:查尔酮异构酶;F3H:黄烷酮3-羟化酶;F3'H:类黄酮 3'-羟化酶;F3'5'H:黄烷酮3' 5'-羟化酶;DFR:二氢黄酮醇还原酶;ANS:花色素合成酶; UFGT:类黄酮3-O-糖苷转移酶
Fig. 2 Anthocyanin biosynthesis pathway PAL: Phenylalanin ammonialyase; C4H: cinnamate 4-hydroxylase; 4CL: 4-coumarate CoA ligase; CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavanone 3-hydroxylase; F3' H: flavonoid 3'-hydroxylase; F3'5'H: flavonoid 3' 5'-hydroxylase; DFR: dihydroflavonol reductase; ANS: anthocyanidin synthase; UFGT: flavonoid 3-O-glucosyltransferase
| 基因名Gene name | 染色体位置Chromosome position | 名称Name | ID | 参考文献Reference |
|---|---|---|---|---|
| c2(ZmCHS) | 4 | 查尔酮合酶Chalcone synthase | Zm00001d052673 | [ |
| chi1(ZmCHI) | 1 | 查尔酮异构酶Chalcone isomerase | Zm00001d034635 | [ |
| fht1(ZmF3H) | 2 | 黄烷酮3-羟化酶Flavanon -3-hydroxylase | Zm00001d001960 | [ |
| pr1(ZmF3'H) | 5 | 黄烷酮3'-羟化酶Flavonoid 3'-hydroxylase | Zm00001d017077 | [ |
| A1(ZmDFR) | 3 | 二氢黄酮醇还原酶Dihydroflavonol 4-reductase | Zm00001d044122 | [ |
| A2(ZmANS) | 5 | 花青素合酶Anthocyanidin aynthase | Zm00001d014914 | [ |
| bz1(ZmUFGT) | 9 | 类黄酮3-O葡萄糖基转移酶Flavonoid 3-Oglucosyltransferase | Zm00001d045055 | [ |
表1 玉米中花青素合成结构基因
Table 1 Genes for anthocyanin synthesis structure in maize
| 基因名Gene name | 染色体位置Chromosome position | 名称Name | ID | 参考文献Reference |
|---|---|---|---|---|
| c2(ZmCHS) | 4 | 查尔酮合酶Chalcone synthase | Zm00001d052673 | [ |
| chi1(ZmCHI) | 1 | 查尔酮异构酶Chalcone isomerase | Zm00001d034635 | [ |
| fht1(ZmF3H) | 2 | 黄烷酮3-羟化酶Flavanon -3-hydroxylase | Zm00001d001960 | [ |
| pr1(ZmF3'H) | 5 | 黄烷酮3'-羟化酶Flavonoid 3'-hydroxylase | Zm00001d017077 | [ |
| A1(ZmDFR) | 3 | 二氢黄酮醇还原酶Dihydroflavonol 4-reductase | Zm00001d044122 | [ |
| A2(ZmANS) | 5 | 花青素合酶Anthocyanidin aynthase | Zm00001d014914 | [ |
| bz1(ZmUFGT) | 9 | 类黄酮3-O葡萄糖基转移酶Flavonoid 3-Oglucosyltransferase | Zm00001d045055 | [ |
| 转录因子 Transcription factor | 基因名 Gene name | 位置 Chromosome position | 调控着色部位 Regulatory staining site | ID | 参考文献 Reference |
|---|---|---|---|---|---|
| MYB | Colored aleurone1 (C1) | 9 | 种子糊粉层 | Zm00001d044975 Zm00001d008695 | [ |
| Purple plant (pl) | 6 | 花和营养组织 | Zm00001d037118 | [ | |
| Pericarp color 1 (p1) | 1 | 种皮、玉米棒和花丝 | Zm00001d028850 | [ | |
| Pericarp color 2 (p2) | 1 | 花药、花丝 | Zm00001d028842 | [ | |
| bHLH | Seed colorcomponent at R1 (r1) | 10 | 种子糊粉层 | Zm00001d026147 | [ |
| Colored plant1 (B1) | 2 | 叶、叶鞘、雄穗、花药 | Zm00001d000236 | [ | |
| WD40 | Pale aleurone color1(pac1) | 5 | 整株植株 | Zm00001eb250750 | [ |
表2 玉米中花青素合成调控基因
Table 2 Genes regulating anthocyanin synthesis in maize
| 转录因子 Transcription factor | 基因名 Gene name | 位置 Chromosome position | 调控着色部位 Regulatory staining site | ID | 参考文献 Reference |
|---|---|---|---|---|---|
| MYB | Colored aleurone1 (C1) | 9 | 种子糊粉层 | Zm00001d044975 Zm00001d008695 | [ |
| Purple plant (pl) | 6 | 花和营养组织 | Zm00001d037118 | [ | |
| Pericarp color 1 (p1) | 1 | 种皮、玉米棒和花丝 | Zm00001d028850 | [ | |
| Pericarp color 2 (p2) | 1 | 花药、花丝 | Zm00001d028842 | [ | |
| bHLH | Seed colorcomponent at R1 (r1) | 10 | 种子糊粉层 | Zm00001d026147 | [ |
| Colored plant1 (B1) | 2 | 叶、叶鞘、雄穗、花药 | Zm00001d000236 | [ | |
| WD40 | Pale aleurone color1(pac1) | 5 | 整株植株 | Zm00001eb250750 | [ |
| [1] |
Dooner HK, Robbins TP, Jorgensen RA. Genetic and developmental control of anthocyanin biosynthesis[J]. Annu Rev Genet, 1991, 25: 173-199.
pmid: 1839877 |
| [2] | Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis[J]. Plant Cell, 1995, 7(7): 1071-1083. |
| [3] | Rhoades MM. The effect of the bronze locus on anthocyanin formation in maize[J]. Am Nat, 1952, 86(827): 105-108. |
| [4] |
Cone KC, Burr FA, Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1[J]. Proc Natl Acad Sci USA, 1986, 83(24): 9631-9635.
doi: 10.1073/pnas.83.24.9631 pmid: 3025847 |
| [5] | Bars-Cortina D, Sakhawat A, Piñol-Felis C, et al. Chemopreventive effects of anthocyanins on colorectal and breast cancer: a review[J]. Semin Cancer Biol, 2022, 81: 241-258. |
| [6] | Dong YH, Wu X, Han L, et al. The potential roles of dietary anthocyanins in inhibiting vascular endothelial cell senescence and preventing cardiovascular diseases[J]. Nutrients, 2022, 14(14): 2836. |
| [7] | Aboonabi A, Singh I, Rose’ Meyer R. Cytoprotective effects of berry anthocyanins against induced oxidative stress and inflammation in primary human diabetic aortic endothelial cells[J]. Chem Biol Interact, 2020, 317: 108940. |
| [8] | Chatham LA, Juvik JA. Linking anthocyanin diversity, hue, and genetics in purple corn[J]. G3, 2021, 11(2): jkaa062. |
| [9] | Paulsmeyer MN, Vermillion KE, Juvik JA. Assessing the diversity of anthocyanin composition in various tissues of purple corn(Zea mays L.)[J]. Phytochemistry, 2022, 201: 113263. |
| [10] |
Taylor LP, Briggs WR. Genetic regulation and photocontrol of anthocyanin accumulation in maize seedlings[J]. Plant Cell, 1990, 2(2): 115-127.
pmid: 2136630 |
| [11] |
Lepiniec L, Debeaujon I, Routaboul JM, et al. Genetics and biochemistry of seed flavonoids[J]. Annu Rev Plant Biol, 2006, 57: 405-430.
pmid: 16669768 |
| [12] |
Alfenito MR, Souer E, Goodman CD, et al. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases[J]. Plant Cell, 1998, 10(7): 1135-1149.
doi: 10.1105/tpc.10.7.1135 pmid: 9668133 |
| [13] | Cai TY, Ge-Zhang SJ, Song MB. Anthocyanins in metabolites of purple corn[J]. Front Plant Sci, 2023, 14: 1154535. |
| [14] |
Springob K, Nakajima JI, Yamazaki M, et al. Recent advances in the biosynthesis and accumulation of anthocyanins[J]. Nat Prod Rep, 2003, 20(3): 288-303.
doi: 10.1039/b109542k pmid: 12828368 |
| [15] |
Franken P, Niesbach-Klösgen U, Weydemann U, et al. The duplicated chalcone synthase genes C2 and Whp(white pollen)of Zea mays are independently regulated; evidence for translational control of Whp expression by the anthocyanin intensifying gene in[J]. EMBO J, 1991, 10(9): 2605-2612.
doi: 10.1002/j.1460-2075.1991.tb07802.x pmid: 1714383 |
| [16] | Li TC, Wang YT, Dong Q, et al. Weighted gene co-expression network analysis reveals key module and hub genes associated with the anthocyanin biosynthesis in maize pericarp[J]. Front Plant Sci, 2022, 13: 1013412. |
| [17] | Han YH, Ding T, Su B, et al. Genome-wide identification, characterization and expression analysis of the Chalcone synthase family in maize[J]. Int J Mol Sci, 2016, 17(2): 161. |
| [18] | Grotewold E, Peterson T. Isolation and characterization of a maize gene encoding chalcone flavonone isomerase[J]. Mol Gen Genet MGG, 1994, 242(1): 1-8. |
| [19] |
Deboo GB, Albertsen MC, Taylor LP. Flavanone 3-hydroxylase transcripts and flavonol accumulation are temporally coordinate in maize anthers[J]. Plant J, 1995, 7(5): 703-713.
doi: 10.1046/j.1365-313x.1995.07050703.x pmid: 7773305 |
| [20] |
Larson R, Bussard JB, Jr Coe EH. Gene-dependent flavonoid 3'-hydroxylation in maize[J]. Biochem Genet, 1986, 24(7-8): 615-624.
pmid: 3753432 |
| [21] |
Sharma M, Chai CL, Morohashi K, et al. Expression of flavonoid 3'-hydroxylase is controlled by P1, the regulator of 3-deoxyflavonoid biosynthesis in maize[J]. BMC Plant Biol, 2012, 12: 196.
doi: 10.1186/1471-2229-12-196 pmid: 23113982 |
| [22] | Sharma M, Cortes-Cruz M, Ahern KR, et al. Identification of the Pr1 gene product completes the anthocyanin biosynthesis pathway of maize[J]. Genetics, 2011, 188(1): 69-79. |
| [23] |
Bernhardt J, Stich K, Schwarz-Sommer Z, et al. Molecular analysis of a second functional A1 gene(dihydroflavonol 4-reductase)in Zea mays[J]. Plant J, 1998, 14(4): 483-488.
pmid: 9670563 |
| [24] | Anirban A, Hayward A, Hong HT, et al. Breaking the tight genetic linkage between the a1 and sh2 genes led to the development of anthocyanin-rich purple-pericarp super-sweetcorn[J]. Sci Rep, 2023, 13(1): 1050. |
| [25] |
Weiss D, van der Luit AH, Kroon JTM, et al. The petunia homologue of the Antirrhinum majus candi and Zea mays A2 flavonoid genes; homology to flavanone 3-hydroxylase and ethylene-forming enzyme[J]. Plant Mol Biol, 1993, 22(5): 893-897.
pmid: 8358035 |
| [26] |
Lesnick ML, Chandler VL. Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters[J]. Plant Physiol, 1998, 117(2): 437-445.
doi: 10.1104/pp.117.2.437 pmid: 9625696 |
| [27] | Wang J, Li DL, Peng YX, et al. The anthocyanin accumulation related ZmBZ1, facilitates seedling salinity stress tolerance via ROS scavenging[J]. Int J Mol Sci, 2022, 23(24): 16123. |
| [28] |
Roth BA, Goff SA, Klein TM, et al. C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites[J]. Plant Cell, 1991, 3(3): 317-325.
pmid: 1840914 |
| [29] |
Reddy AR, Scheffler B, Madhuri G, et al. Chalcone synthase in rice(Oryza sativa L.): detection of the CHS protein in seedlings and molecular mapping of the chs locus[J]. Plant Mol Biol, 1996, 32(4): 735-743.
pmid: 8980525 |
| [30] | Jiang WB, Yin QG, Wu RR, et al. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana[J]. J Exp Bot, 2015, 66(22): 7165-7179. |
| [31] | Chao N, Wang RF, Hou C, et al. Functional characterization of two chalcone isomerase(CHI)revealing their responsibility for anthocyanins accumulation in mulberry[J]. Plant Physiol Biochem, 2021, 161: 65-73. |
| [32] | 麻新妍, 南春利, 薛永常. 植物查尔酮异构酶的结构与功能研究概述[J]. 生物学教学, 2022, 47(1): 2-4. |
| Ma XY, Nan CL, Xue YC. Review on the structure and function of plant Chalcone isomerase[J]. Biol Teach, 2022, 47(1): 2-4. | |
| [33] |
段玥彤, 王鹏年, 张春宝, 等. 植物黄烷酮-3-羟化酶基因研究进展[J]. 生物技术通报, 2022, 38(6): 27-33.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1253 |
| Duan YT, Wang PN, Zhang CB, et al. Research progress in plant flavanone-3-hydroxylase gene[J]. Biotechnol Bull, 2022, 38(6): 27-33. | |
| [34] |
O'Reilly C, Shepherd NS, Pereira A, et al. Molecular cloning of the a1 locus of Zea mays using the transposable elements En and Mu1[J]. EMBO J, 1985, 4(4): 877-882.
doi: 10.1002/j.1460-2075.1985.tb03713.x pmid: 15938046 |
| [35] | Haselmair-Gosch C, Miosic S, Nitarska D, et al. Great cause-small effect: undeclared genetically engineered orange petunias harbor an inefficient dihydroflavonol 4-reductase[J]. Front Plant Sci, 2018,9: 149. |
| [36] |
Klein TM, Roth BA, Fromm ME. Regulation of anthocyanin biosynthetic genes introduced into intact maize tissues by microprojectiles[J]. Proc Natl Acad Sci USA, 1989, 86(17): 6681-6685.
doi: 10.1073/pnas.86.17.6681 pmid: 16594066 |
| [37] |
Nakajima J, Tanaka Y, Yamazaki M, et al. Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis[J]. J Biol Chem, 2001, 276(28): 25797-25803.
doi: 10.1074/jbc.M100744200 pmid: 11316805 |
| [38] |
Reddy AM, Reddy VS, Scheffler BE, et al. Novel transgenic rice overexpressing anthocyanidin synthase accumulates a mixture of flavonoids leading to an increased antioxidant potential[J]. Metab Eng, 2007, 9(1): 95-111.
pmid: 17157544 |
| [39] |
Piazza P, Procissi A, Jenkins GI, et al. Members of the c1/pl1 regulatory gene family mediate the response of maize aleurone and mesocotyl to different light qualities and cytokinins[J]. Plant Physiol, 2002, 128(3): 1077-1086.
pmid: 11891262 |
| [40] | Riaz B, Chen HQ, Wang J, et al. Overexpression of maize ZmC1 and ZmR transcription factors in wheat regulates anthocyanin biosynthesis in a tissue-specific manner[J]. Int J Mol Sci, 2019, 20(22): 5806. |
| [41] | Li TC, Zhang W, Yang HY, et al. Comparative transcriptome analysis reveals differentially expressed genes related to the tissue-specific accumulation of anthocyanins in pericarp and aleurone layer for maize[J]. Sci Rep, 2019, 9(1): 2485. |
| [42] |
Zhang P, Chopra S, Peterson T. A segmental gene duplication generated differentially expressed myb-homologous genes in maize[J]. Plant Cell, 2000, 12(12): 2311-2322.
doi: 10.1105/tpc.12.12.2311 pmid: 11148280 |
| [43] | Luo MJ, Lu BS, Shi YX, et al. A newly characterized allele of ZmR1 increases anthocyanin content in whole maize plant and the regulation mechanism of different ZmR1 alleles[J]. Theor Appl Genet, 2022, 135(9): 3039-3055. |
| [44] | Radicella JP, Brown D, Tolar LA, et al. Allelic diversity of the maize B regulatory gene: different leader and promoter sequences of two B alleles determine distinct tissue specificities of anthocyanin production[J]. Genes Dev, 1992, 6(11): 2152-2164. |
| [45] |
Selinger DA, Chandler VL. A mutation in the pale aleurone color1 gene identifies a novel regulator of the maize anthocyanin pathway[J]. Plant Cell, 1999, 11(1): 5-14.
pmid: 9878628 |
| [46] | Yan HL, Pei XN, Zhang H, et al. MYB-mediated regulation of anthocyanin biosynthesis[J]. Int J Mol Sci, 2021, 22(6): 3103. |
| [47] |
Du H, Wang YB, Xie Y, et al. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants[J]. DNA Res, 2013, 20(5): 437-448.
doi: 10.1093/dnares/dst021 pmid: 23690543 |
| [48] | Zhang YZ, Xu SZ, Ma HP, et al. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers[J]. Plant Physiol Biochem, 2021, 164: 279-288. |
| [49] | Wang L, Deng RY, Bai YC, et al. Tartary buckwheat R2R3-MYB gene FtMYB3 negatively regulates anthocyanin and proanthocyanin biosynthesis[J]. Int J Mol Sci, 2022, 23(5): 2775. |
| [50] | Paulsmeyer MN, Juvik JA. R3-MYB repressor Mybr97 is a candidate gene associated with the Anthocyanin3 locus and enhanced anthocyanin accumulation in maize[J]. Theor Appl Genet, 2023, 136(3): 55. |
| [51] | Li Y, Fang XJ, Lin ZW. Convergent loss of anthocyanin pigments is controlled by the same MYB gene in cereals[J]. J Exp Bot, 2022, 73(18): 6089-6102. |
| [52] |
Cone KC, Cocciolone SM, Burr FA, et al. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant[J]. Plant Cell, 1993, 5(12): 1795-1805.
pmid: 8305872 |
| [53] | Qian YC, Zhang TY, Yu Y, et al. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses[J]. Front Plant Sci, 2021, 12: 677611. |
| [54] | Gao QQ, Song WL, Li X, et al. Genome-wide identification of bHLH transcription factors: discovery of a candidate regulator related to flavonoid biosynthesis in Erigeron breviscapus[J]. Front Plant Sci, 2022, 13: 977649. |
| [55] |
Robbins MP, Paolocci F, Hughes JW, et al. Sn, a maize bHLH gene, modulates anthocyanin and condensed tannin pathways in Lotus corniculatus[J]. J Exp Bot, 2003, 54(381): 239-248.
doi: 10.1093/jxb/erg022 pmid: 12493851 |
| [56] |
Jain BP, Pandey S. WD40 repeat proteins: signalling scaffold with diverse functions[J]. Protein J, 2018, 37(5): 391-406.
doi: 10.1007/s10930-018-9785-7 pmid: 30069656 |
| [57] | Chen L, Cui YM, Yao YH, et al. Genome-wide identification of WD40 transcription factors and their regulation of the MYB-bHLH-WD40(MBW)complex related to anthocyanin synthesis in Qingke(Hordeum vulgare L. var. nudum Hook. f.)[J]. BMC Genomics, 2023, 24(1): 166. |
| [58] | Wang XC, Wu J, Guan ML, et al. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis[J]. Plant J, 2020, 101(3): 637-652. |
| [59] | Li YQ, Shan XT, Gao RF, et al. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida[J]. Commun Biol, 2020, 3(1): 396. |
| [60] | Zhang LX, Wang YP, Yue ML, et al. FaMYB 5 interacts with FaBBX24 to regulate anthocyanin and proanthocyanidin biosynthesis in strawberry(Fragaria × ananassa)[J]. Int J Mol Sci, 2023, 24(15): 12185. |
| [61] | Verweij W, Spelt CE, Bliek M, et al. Functionally similar WRKY proteins regulate vacuolar acidification in Petunia and hair development in Arabidopsis[J]. Plant Cell, 2016, 28(3): 786-803. |
| [62] | Amato A, Cavallini E, Walker AR, et al. The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine[J]. Plant J, 2019, 99(6): 1220-1241. |
| [63] | Tang Y, Guo JF, Zhang TT, et al. Genome-wide analysis of WRKY gene family and the dynamic responses of key WRKY genes involved in Ostrinia furnacalis attack in Zea mays[J]. Int J Mol Sci, 2021, 22(23): 13045. |
| [64] |
Zhang SY, Chen YX, Zhao LL, et al. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10[J]. Tree Physiol, 2020, 40(3): 413-423.
doi: 10.1093/treephys/tpaa004 pmid: 32031661 |
| [65] | Fang X, Zhang LZ, Wang LJ. The transcription factor MdERF78 is involved in ALA-induced anthocyanin accumulation in apples[J]. Front Plant Sci, 2022, 13: 915197. |
| [66] | An JP, Yao JF, Xu RR, et al. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation[J]. Plant Cell Environ, 2018, 41(11): 2678-2692. |
| [67] |
Liu XQ, Li SZ, Yang WZ, et al. Synthesis of seed-specific bidirectional promoters for metabolic engineering of anthocyanin-rich maize[J]. Plant Cell Physiol, 2018, 59(10): 1942-1955.
doi: 10.1093/pcp/pcy110 pmid: 29917151 |
| [68] | Liu XQ, Yang WZ, Mu BN, et al. Engineering of ‘Purple Embryo Maize’ with a multigene expression system derived from a bidirectional promoter and self-cleaving 2A peptides[J]. Plant Biotechnol J, 2018, 16(6): 1107-1109. |
| [69] | Wang J, Yuan ZP, Li DL, et al. Transcriptome analysis revealed the potential molecular mechanism of anthocyanidins’ improved salt tolerance in maize seedlings[J]. Plants, 2023, 12(15): 2793. |
| [70] | Johnson ET, Berhow MA, Dowd PF. Constitutive expression of the maize genes B1 and C1 in transgenic Hi II maize results in differential tissue pigmentation and generates resistance to Helicoverpa zea[J]. J Agric Food Chem, 2010, 58(4): 2403-2409. |
| [71] | Nanda DK, Chase SS. An embryo marker for detecting monoploids of maize(Zea mays L.)[J]. Crop Sci, 1966, 6(2): 213-215. |
| [1] | 王秋月, 段鹏亮, 李海笑, 刘宁, 曹志艳, 董金皋. 玉米大斑病菌cDNA文库的构建及转录因子StMR1互作蛋白的筛选[J]. 生物技术通报, 2024, 40(6): 281-289. |
| [2] | 张美玉, 赵玉斌, 王灵云, 宋元达, 赵新河, 任晓洁. 微藻破囊壶菌产功能性脂肪酸DHA研究进展[J]. 生物技术通报, 2024, 40(6): 81-94. |
| [3] | 吴泽航, 杨中义, 鄢毅铖, 贾永红, 吴月燕, 谢晓鸿. 比利时杜鹃花类黄酮3'-羟化酶(F3'H)基因克隆及功能分析[J]. 生物技术通报, 2024, 40(6): 251-259. |
| [4] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [5] | 孙亚楠, 王春雪, 王欣, 杜秉海, 刘凯, 汪城墙. 萎缩芽孢杆菌CNY01的生防特性及其对玉米的抗盐促生作用[J]. 生物技术通报, 2024, 40(5): 248-260. |
| [6] | 王佳玮, 李晨, 刘建利, 周世杰, 易嘉敏, 杨谨源, 康鹏. 内生真菌接种方式对青贮玉米幼苗生长的影响[J]. 生物技术通报, 2024, 40(4): 189-202. |
| [7] | 胡伊娃, 陈露. 玉米野生种基因组研究进展及应用[J]. 生物技术通报, 2024, 40(3): 14-24. |
| [8] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [9] | 王俊芳, 黄秋斌, 张飘丹, 张彭湃. Surfactin的结构、生物合成及其在生物防治中的作用[J]. 生物技术通报, 2024, 40(1): 100-112. |
| [10] | 常泸尹, 王中华, 李凤敏, 高梓源, 张辉红, 王祎, 李芳, 韩燕来, 姜瑛. 玉米根际多功能促生菌的筛选及其对冬小麦-夏玉米轮作体系产量提升效果[J]. 生物技术通报, 2024, 40(1): 231-242. |
| [11] | 陈治民, 李翠, 韦继天, 李昕然, 刘峄, 郭强. 绿原酸生物合成调控及其应用研究进展[J]. 生物技术通报, 2024, 40(1): 57-71. |
| [12] | 李亮, 徐姗姗, 姜艳军. 生物合成法生产麦角硫因的研究进展[J]. 生物技术通报, 2024, 40(1): 86-99. |
| [13] | 王宝宝, 王海洋. 理想株型塑造之于玉米耐密改良[J]. 生物技术通报, 2023, 39(8): 11-30. |
| [14] | 张道磊, 甘雨军, 乐亮, 普莉. 玉米产量性状的表观遗传调控机制和育种应用[J]. 生物技术通报, 2023, 39(8): 31-42. |
| [15] | 冷燕, 马晓薇, 陈光, 任鹤, 李翔. 玉米高产竞赛助力中国玉米种业振兴[J]. 生物技术通报, 2023, 39(8): 4-10. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||