








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 161-171.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1153
刘蓉1,2( ), 田闵玉1,2, 李光泽1,2, 谭成方2, 阮颖2, 刘春林1,2(
), 田闵玉1,2, 李光泽1,2, 谭成方2, 阮颖2, 刘春林1,2( )
)
收稿日期:2023-12-08
出版日期:2024-06-26
发布日期:2024-05-23
通讯作者:
刘春林,男,博士,教授,研究方向:油菜遗传育种;E-mail: liucl@hunau.edu.cn作者简介:刘蓉,女,硕士,研究方向:油菜遗传育种;E-mail: 2804035191@qq.com
基金资助:
LIU Rong1,2( ), TIAN Min-yu1,2, LI Guang-ze1,2, TAN Cheng-fang2, RUAN Ying2, LIU Chun-lin1,2(
), TIAN Min-yu1,2, LI Guang-ze1,2, TAN Cheng-fang2, RUAN Ying2, LIU Chun-lin1,2( )
)
Received:2023-12-08
Published:2024-06-26
Online:2024-05-23
摘要:
【目的】 REVEILLE家族基因具有重要生物学功能,而在甘蓝型油菜(Brassica napus L.)中该家族基因研究较少,深入认识甘蓝型油菜中RVE家族基因功能及其与非生物胁迫的关系。【方法】 利用生物信息学方法,在甘蓝型油菜、白菜(Brassica rapa)和甘蓝(B. oleracea)中分别鉴定到33、16和16个RVE基因,并对其系统进化、染色体定位、基因结构和理化性质以及启动子顺式元件进行分析。【结果】 所有的RVE蛋白被分为两个亚家族。33个BnaRVE分别定位在18条A亚基因组的染色体和15条C亚基因组的染色体上。大部分成员属于较稳定的疏水蛋白;多数Ka/Ks值小于1,说明该家族受到强烈的纯化选择作用。基因结构变异较大,内含子数目在4-11之间。共线性分析结果表明,甘蓝型油菜和拟南芥、白菜、甘蓝存在大量的同源基因。大部分甘蓝型油菜RVE家族成员在子叶和叶都有表达且表达量最多。BnaRVE启动子上含有大量与激素和生物逆境相关的元件,通过定量PCR分析,4个BnaRVE在ABA与MeJA诱导和低温胁迫下的表达量显著上调。【结论】 在甘蓝型油菜基因组中鉴定出33个BnaRVE家族成员。不同基因在不同发育时期和不同组织中表现出不同的表达模式。不同基因对各种胁迫存在响应,且表达模式不一。RVE家族成员对ABA、MeJA和冷胁迫具有正向响应功能。
刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171.
LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L.[J]. Biotechnology Bulletin, 2024, 40(6): 161-171.
| Gene name | Forword primer sequence(5'-3') | Reverse primer sequence(5'-3') |
|---|---|---|
| BnaRVE1d | TGTCTCCTGTTTCCTTTGCATCACC | AACTCTCTCTGTTTACGAGTCTCTCTCT |
| BnaRVE8b | TTCGACCTCCGGTATGGGAAG | GAATTACCTTGAAGCACCGGAGG |
| BnaRVE3c | GTGTGGCTCTTACAACTTCAAACGCAT | CACCCAATTCCTCTTTGATCACTGGAA |
| BnaRVE5b BnaActin2 | TCTCCACTTATTCGATCGAGATTGGAAG GTCTTCTCTGCTCTTCTCA | GTTCGTTAGCACCGCTCTTCTG CCATTCCAGTTCCATTGTC |
表1 甘蓝型油菜RVE基因家族表达分析的实时荧光定量引物
Table 1 Real-time fluorescent quantitative primers for RVE gene family expression analysis in B. napus
| Gene name | Forword primer sequence(5'-3') | Reverse primer sequence(5'-3') |
|---|---|---|
| BnaRVE1d | TGTCTCCTGTTTCCTTTGCATCACC | AACTCTCTCTGTTTACGAGTCTCTCTCT |
| BnaRVE8b | TTCGACCTCCGGTATGGGAAG | GAATTACCTTGAAGCACCGGAGG |
| BnaRVE3c | GTGTGGCTCTTACAACTTCAAACGCAT | CACCCAATTCCTCTTTGATCACTGGAA |
| BnaRVE5b BnaActin2 | TCTCCACTTATTCGATCGAGATTGGAAG GTCTTCTCTGCTCTTCTCA | GTTCGTTAGCACCGCTCTTCTG CCATTCCAGTTCCATTGTC |

图4 甘蓝型油菜RVE家族成员启动子区域结合的顺式作用元件(A)及前10位CRE在基因组上的分布(B)
Fig. 4 Genomic distribution of cis-acting elements binding to promoter region of RVE family members in B. napus(A)and the top 10 CRE(B)in genome

图5 甘蓝型油菜RVE家族种内共线性分析(A)及RVE家族基因在甘蓝型油菜、拟南芥、甘蓝和白菜之间的共线性分析(B-D)
Fig. 5 Intra-specic collinearity analysis of B. napus RVE family(A)and collinearity analysis of RVE family genes among A. thaliana, B. napus, B. rapa and B. oleracea(B-D)
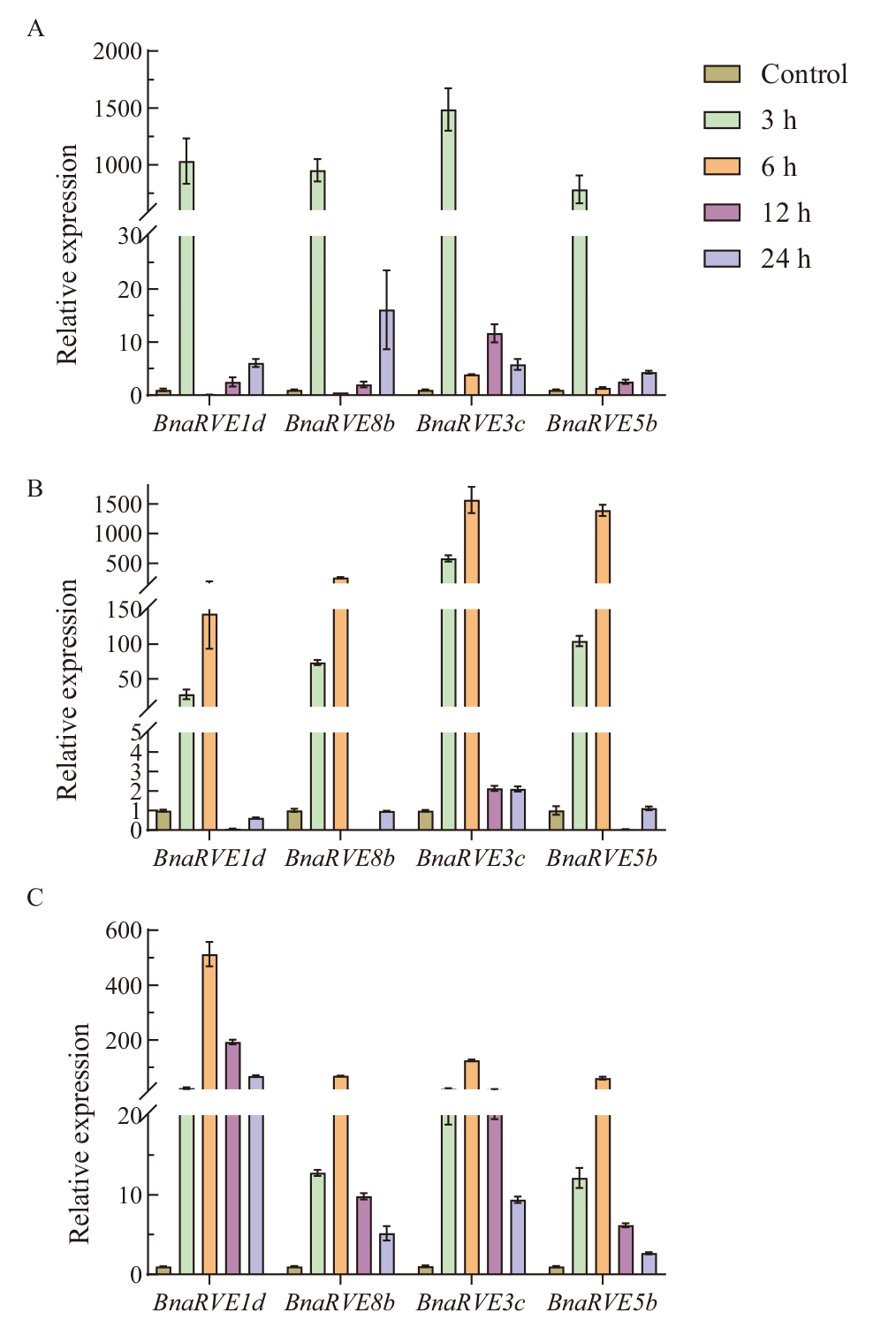
图7 BnaRVE在不同胁迫处理下的RT-qPCR表达分析 A:BnaRVE在MeJA处理中的表达;B:BnaRVE在ABA处理中的表达;C:BnaRVE在冷处理中的表达
Fig. 7 RT-qPCR expression analysis of BnaRVE under different stress treatments A: Expression of BnaRVE in MeJA processing.B: Expression of BnaRVE in ABA processing. C: Expression of BnaRVE in cold treatment
| [1] | Wu GF, Cao AH, Wen YH, et al. Characteristics and functions of MYB(v-Myb avivan myoblastsis virus oncogene homolog)-related genes in Arabidopsis thaliana[J]. Genes, 2023, 14(11): 2026. |
| [2] | Garighan J, Dvorak E, Estevan J, et al. The identification of small RNAs differentially expressed in apple buds reveals a potential role of the Mir159-MYB regulatory module during dormancy[J]. Plants, 2021, 10(12): 2665. |
| [3] | Premathilake AT, Ni JB, Bai SL, et al. R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit[J]. Planta, 2020, 252(4): 59. |
| [4] | Zhang CH, Ma RJ, Xu JL, et al. Genome-wide identification and classification of MYB superfamily genes in peach[J]. PLoS One, 2018, 13(6): e0199192. |
| [5] | Liu JX, Wang J, Wang MQ, et al. Genome-wide analysis of the R2R3-MYB gene family in Fragaria × ananassa and its function identification during anthocyanins biosynthesis in pink-flowered strawberry[J]. Front Plant Sci, 2021, 12: 702160. |
| [6] | Li JL, Han GL, Sun CF, et al. Research advances of MYB transcription factors in plant stress resistance and breeding[J]. Plant Signal Behav, 2019, 14(8): 1613131. |
| [7] | Tian YY, Li W, Wang MJ, et al. REVEILLE 7 inhibits the expression of the circadian clock gene EARLY FLOWERING 4 to fine-tune hypocotyl growth in response to warm temperatures[J]. J Integr Plant Biol, 2022, 64(7): 1310-1324. |
| [8] |
Scandola S, Mehta D, Li QM, et al. Multi-omic analysis shows REVEILLE clock genes are involved in carbohydrate metabolism and proteasome function[J]. Plant Physiol, 2022, 190(2): 1005-1023.
doi: 10.1093/plphys/kiac269 pmid: 35670757 |
| [9] | Liu Z, Zhu XX, Liu WJ, et al. Characterization of the REVEILLE family in Rosaceae and role of PbLHY in flowering time regulation[J]. BMC Genomics, 2023, 24(1): 49. |
| [10] | Liu CY, Zhang QQ, Dong J, et al. Genome-wide identification and characterization of mungbean circadian clock associated 1 like genes reveals an important role of vrcca1l26 in flowering time regulation[J]. BMC Genomics, 2022, 23(1): 374. |
| [11] | Creux N, Harmer S. Circadian rhythms in plants[J]. Cold Spring Harb Perspect Biol, 2019, 11(9): a034611. |
| [12] | Shan BH, Wang W, Cao JF, et al. Soybean GmMYB133 inhibits hypocotyl elongation and confers salt tolerance in Arabidopsis[J]. Front Plant Sci, 2021, 12: 764074. |
| [13] | Xue XW, Sun KW, Zhu ZQ. CIRCADIAN CLOCK ASSOCIATED 1 gates morning phased auxin response in Arabidopsis thaliana[J]. Biochem Biophys Res Commun, 2020, 527(4): 935-940. |
| [14] | Muroya M, Oshima H, Kobayashi S, et al. Circadian clock in Arabidopsis thaliana determines flower opening time early in the morning and dominantly closes early in the afternoon[J]. Plant Cell Physiol, 2021, 62(5): 883-893. |
| [15] |
Gray JA, Shalit-Kaneh A, Chu DN, et al. The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size[J]. Plant Physiol, 2017, 173(4): 2308-2322.
doi: 10.1104/pp.17.00109 pmid: 28254761 |
| [16] | Yang LW, Jiang ZM, Jing YJ, et al. PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis[J]. J Integr Plant Biol, 2020, 62(9): 1372-1384. |
| [17] | Liu DG, Tang D, Xie M, et al. Agave REVEILLE1 regulates the onset and release of seasonal dormancy in Populus[J]. Plant Physiol, 2023, 191(3): 1492-1504. |
| [18] | Zhang Y, Chen CQ, Cui YL, et al. Potential regulatory genes of light induced anthocyanin accumulation in sweet cherry identified by combining transcriptome and metabolome analysis[J]. Front Plant Sci, 2023, 14: 1238624. |
| [19] | Bian SM, Li RH, Xia SQ, et al. Soybean CCA1-like MYB transcription factor GmMYB133 modulates isoflavonoid biosynthesis[J]. Biochem Biophys Res Commun, 2018, 507(1/2/3/4): 324-329. |
| [20] | Li XY, Wu T, Liu HT, et al. REVEILLE transcription factors contribute to the nighttime accumulation of anthocyanins in ‘red zaosu’(Pyrus bretschneideri rehd.) pear fruit skin[J]. Int J Mol Sci, 2020, 21(5): 1634. |
| [21] | Abuelsoud W, Cortleven A, Schmülling T. Photoperiod stress induces an oxidative burst-like response and is associated with increased apoplastic peroxidase and decreased catalase activities[J]. J Plant Physiol, 2020, 253: 153252. |
| [22] | Li BJ, Gao ZH, Liu XY, et al. Transcriptional profiling reveals a time-of-day-specific role of REVEILLE 4/8 in regulating the first wave of heat shock-induced gene expression in Arabidopsis[J]. Plant Cell, 2019, 31(10): 2353-2369. |
| [23] | Kidokoro S, Hayashi K, Haraguchi H, et al. Posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in Arabidopsis[J]. Proc Natl Acad Sci USA, 2021, 118(10): e2021048118. |
| [24] |
Chen S, Huang HA, Chen JH, et al. SgRVE6, a LHY-CCA1-like transcription factor from fine-stem stylo, upregulates NB-LRR gene expression and enhances cold tolerance in tobacco[J]. Front Plant Sci, 2020, 11: 1276.
doi: 10.3389/fpls.2020.01276 pmid: 32973836 |
| [25] |
李利霞, 陈碧云, 闫贵欣, 等. 中国油菜种质资源研究利用策略与进展[J]. 植物遗传资源学报, 2020, 21(1): 1-19.
doi: 10.13430/j.cnki.jpgr.20200109005 |
| Li LX, Chen BY, Yan GX, et al. Proposed strategies and current progress of research and utilization of oilseed rape germplasm in China[J]. J Plant Genet Resour, 2020, 21(1): 1-19. | |
| [26] |
廖琼, 刘佳林, 刘婧, 等. 油菜ABCC家族基因生物信息学分析及其对镉胁迫的转录响应[J]. 中国油料作物学报, 2023, 45(2): 271-282.
doi: 10.19802/j.issn.1007-9084.2022045 |
| Liao Q, Liu JL, Liu J, et al. Bioinformatics of ABCC family genes and their transcriptional response to cadmium stress in Brassica napus L[J]. Chin J Oil Crop Sci, 2023, 45(2): 271-282. | |
| [27] | Subramanian B, Gao SH, Lercher MJ, et al. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees[J]. Nucleic Acids Res, 2019, 47(W1): W270-W275. |
| [28] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining[J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [29] | Sukumaran S, Lethin J, Liu X, et al. Genome-wide analysis of MYB transcription factors in the wheat genome and their roles in salt stress response[J]. Cells, 2023, 12(10): 1431. |
| [30] | Haxim Y, Kahar G, Zhang XC, et al. Genome-wide characterization of the chitinase gene family in wild apple(Malus sieversii)and domesticated apple(Malus domestica)reveals its role in resistance to Valsa mali[J]. Front Plant Sci, 2022, 13: 1007936. |
| [31] | Kidokoro S, Konoura I, Soma F, et al. Clock-regulated coactivators selectively control gene expression in response to different temperature stress conditions in Arabidopsis[J]. Proc Natl Acad Sci USA, 2023, 120(16): e2216183120. |
| [32] | Chen YS, Chao YC, Tseng TW, et al. Two MYB-related transcription factors play opposite roles in sugar signaling in Arabidopsis[J]. Plant Mol Biol, 2017, 93(3): 299-311. |
| [33] |
Wang K, Bu TT, Cheng Q, et al. Two homologous LHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses[J]. New Phytol, 2021, 229(5): 2660-2675.
doi: 10.1111/nph.17019 pmid: 33095906 |
| [1] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [2] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [3] | 李博静, 郑腊梅, 吴乌云, 高飞, 周宜君. 西蒙得木HSP20基因家族的进化、表达和功能分析[J]. 生物技术通报, 2024, 40(6): 190-202. |
| [4] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [5] | 苑海鹏, 叶云舒, 司皓, 纪秋研, 张玉红. 丛枝菌根真菌对植物逆境胁迫抗性及次生代谢产物合成的影响[J]. 生物技术通报, 2024, 40(6): 45-56. |
| [6] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [7] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [8] | 娄银, 高浩竣, 王茜, 牛景萍, 王敏, 杜维俊, 岳爱琴. 大豆GmHMGS基因的鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(4): 110-121. |
| [9] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [10] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [11] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [12] | 杨伟成, 孙岩, 杨倩, 王壮琳, 马菊花, 薛金爱, 李润植. 陆地棉FAX家族的全基因组鉴定及GhFAX1的功能分析[J]. 生物技术通报, 2024, 40(3): 155-169. |
| [13] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [14] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [15] | 李亚男, 张豪杰, 梁梦静, 罗涛, 李旺宁, 张春辉, 季春丽, 李润植, 薛金爱, 崔红利. 雨生红球藻钙依赖蛋白激酶(CDPK)家族鉴定与表达分析[J]. 生物技术通报, 2024, 40(2): 300-312. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||