








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 122-138.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0410
冯凯月1( ), 赵鑫焱1, 李子妍1, 邱江明2(
), 赵鑫焱1, 李子妍1, 邱江明2( ), 曹一博1(
), 曹一博1( )
)
收稿日期:2024-04-29
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
邱江明,男,硕士,副教授,研究方向:经济林种苗繁育及经济林林下复合经营;E-mail: 18270730388@163.com;作者简介:冯凯月,女,硕士研究生,研究方向:经济林(果树)培育与应用;E-mail: 2213298711@qq.com
基金资助:
FENG Kai-yue1( ), ZHAO Xin-yan1, LI Zi-yan1, QIU Jiang-ming2(
), ZHAO Xin-yan1, LI Zi-yan1, QIU Jiang-ming2( ), CAO Yi-bo1(
), CAO Yi-bo1( )
)
Received:2024-04-29
Published:2024-10-26
Online:2024-11-20
摘要:
土壤盐碱化是制约全球农业发展的主要环境因素之一。培育耐盐碱作物是应对土壤盐碱化的根本措施,但目前仍面临基因、种质资源匮乏等挑战。挖掘耐盐碱新基因,阐明植物耐盐碱应答的分子机制对培育耐盐碱作物至关重要。盐碱胁迫包括中性盐胁迫和碱性盐胁迫,中性盐胁迫会产生渗透胁迫,Na+、Cl-积累过量还会导致离子毒害,引起氧化胁迫等一系列次生胁迫。与中性盐胁迫相比,碱性盐胁迫还会引起高pH胁迫。论文综述了盐碱胁迫对植物生长发育的影响,总结了近十年植物耐盐碱应答机制的重要研究进展。包括植物对盐胁迫的感知与信号转导,以及植物响应中性盐胁迫和碱性盐胁迫引起的渗透胁迫、离子毒害、氧化胁迫、碳酸氢盐和碳酸盐胁迫、高pH胁迫的分子机制。在此基础上,还讨论了耐盐碱基因在作物育种方面的应用,并提出了提高植物耐盐碱性需要进一步研究的关键科学问题。旨在加深对植物响应盐碱胁迫分子机制的认识,为培育高产、优质的耐盐碱品种及提升盐碱地可开发利用率提供理论基础。
冯凯月, 赵鑫焱, 李子妍, 邱江明, 曹一博. 植物响应盐碱胁迫的分子机制研究进展[J]. 生物技术通报, 2024, 40(10): 122-138.
FENG Kai-yue, ZHAO Xin-yan, LI Zi-yan, QIU Jiang-ming, CAO Yi-bo. Research Advances in the Molecular Mechanisms of Plant Response to Saline-alkali Stress[J]. Biotechnology Bulletin, 2024, 40(10): 122-138.

图1 植物维持离子稳态适应中性盐胁迫的信号通路 盐胁迫下,OSCA1和GIPC分别感知外界渗透压和Na+浓度变化,介导Ca2+内流。SOS3与Ca2+结合后与SOS2互作,激活位于质膜上的SOS1促进Na+转运到胞外,SCaBP8和CBL8发挥类似功能。SOS2与AtANN4互作抑制其介导的Ca2+内流。14-3-3λ/κ蛋白和激酶PKS5抑制SOS2的活性。J3、PKS5和14-3-3ω参与调控AHA2的活性。SCaBP8的磷酸化释放其对AKT1的抑制,促进盐胁迫下K+的吸收。位于液泡膜上的NHXs、V H+-ATP酶、CLCs、ZmMATE29分别参与Na+和Cl-的区隔化。14-3-3蛋白通过CPK21依赖与非依赖途径激活K+外流通道GORK,AtPP2CA抑制GORK的活性
Fig. 1 Signaling pathways for homeostasis adaptation in plants under neutral salt stress Under salt stress, OSCA1 and GIPC sense changes in external osmotic pressure and Na+ concentration, respectively, mediating Ca2+ influx. SOS3 binds to Ca2+ and interacts with SOS2, activating SOS1 located on the plasma membrane to promote Na+ transport to the extracellular cell. SCaBP8 and CBL8 perform similar functions. SOS2 interacts with AtANN4 to inhibit its mediation of Ca2+ influx. The 14-3-3λ/κ protein and kinase PKS5 inhibit the activity of SOS2. J3, PKS5, and 14-3-3ω are involved in regulating the activity of AHA2. The phosphorylation of SCaBP8 releases its inhibition of AKT1, promoting K+ uptake under salt stress. NHXs, V H+-ATPase, CLCs, and ZmMATE29 located on the vacuolar membrane are involved in the compartmentalization of Na+ and Cl-, respectively. The 14-3-3 proteins activate the K+ efflux channel GORK through both CPK21-dependent and independent pathway, while AtPP2CA inhibits the activity of GORK
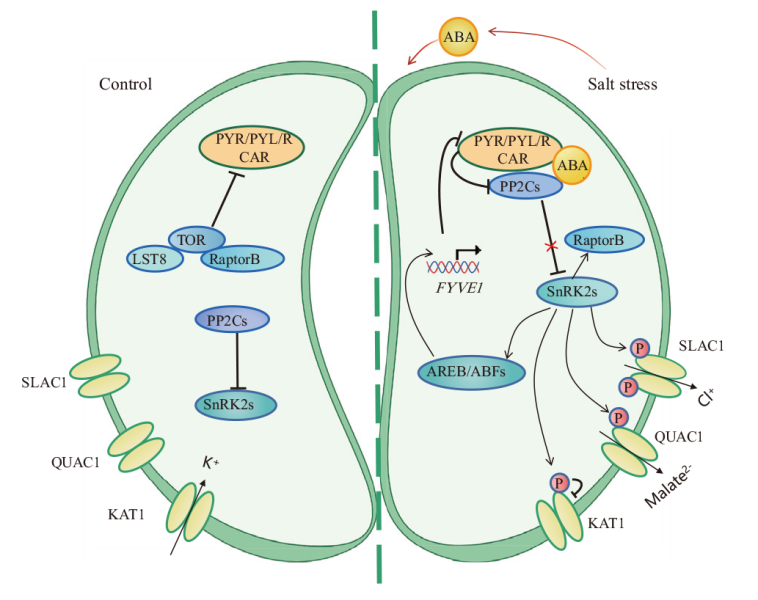
图2 中性盐胁迫下ABA信号介导气孔关闭的分子机制 正常条件下,PP2C与SnRK2s蛋白结合,抑制其活性;TOR磷酸化PYR/PYL/RCAR,抑制PYR/PYL/RCAR与ABA结合。渗透胁迫下,PP2C与PYR/PYL/RCAR形成复合物,释放SnRK2s蛋白,激活转录因子AREB/ABF及阴离子通道SLAC1/QUAC1。拟南芥中ABF4激活FYVE1的表达,促进PYR/PYL降解。SnRK2s磷酸化RaptorB可以促进TOR复合物的解离。SnRK2s还可以抑制KAT1的活性,降低细胞膨压,促进气孔闭合
Fig. 2 Molecular mechanism of ABA signaling mediating stomatal closure under neutral salt stress Under normal conditions, PP2C binds to SnRK2s protein and inhibits its activity; TOR phosphorylates PYR/PYL/RCAR and suppresses PYR/PYL/RCAR binding to ABA. Under osmotic stress, PP2C forms a complex with PYR/PYL/RCAR to release SnRK2s protein and activate the transcription factor AREB/ABF and the anion channel SLAC1/QUAC1. ABF4 in Arabidopsis thaliana L. activates the expressions of FYVE1 and promote PYR/PYL degradation. Phosphorylation of RaptorB by SnRK2s promotes the dissociation of the TOR complex. SnRK2s can also inhibit the activity of KAT1, decrease cellular turgor pressure, and promote stomatal closure

图3 中性盐胁迫下离子转运蛋白介导的Na+、K+、Cl-转运和区隔化 Na+通过NSCCs以及高亲和性钾转运蛋白(如OSHKT2;1)进入根表皮细胞,SOS1介导Na+外排。GORK和NSCCs介导盐胁迫下K+外流。HAK5、AKT1介导K+内流。根皮层细胞中,NPF2.5负责Cl-外排,而ZmMATE29和CLCs将Cl-区隔化到液泡中。NHX可能参与调控Na+区隔化到液泡中。盐胁迫诱导 ZmESBL表达水平增加,促进凯氏带发育,抑制Na+通过质外体途径装载到木质部。CIF-GSO1/SGN3-SGN1同样可以增强凯氏带质外体屏障的功能。木质部薄壁细胞中,OsHKT2;1、SOS1介导Na+装载到木质部;HKT1、ZmHAK4和OsHAK12将木质部汁液中的Na+转运到木薄壁细胞。SKOR介导K+装载到木质部导管中;ZmHKT2将K+从木质部导管中去除。NPF2.4和SLAH1/3介导Cl-在木质部导管的装载和卸载
Fig. 3 Ion transporters mediating the transport and compartmentalization of Na+, K+, and Cl- under neutral salt stress Na+ enters the root epidermal cells through NSCCs and high-affinity K+ transporters such as OSHKT2;1, while SOS1 mediates Na+ efflux. GORK and NSCCs mediate K+ efflux under salt stress. HAK5 and AKT1 mediate K+ influx. In root cortex cells, NPF2.5 is responsible for Cl- efflux, while ZmMATE29 and CLCs transports Cl- into the vacuole. NHX might be involved in the regulation of Na+ compartmentalization into the vacuole. Salt stress induces the expressions of ZmESBL increased, promoting CS development and inhibiting Na+ loading into the xylem via the apoplastic pathway. The CIF-GSO1/SGN3-SGN1 pathway can also enhance the function of the apoplastic barrier. In xylem parenchyma cells, Na+ loading into the xylem is mediated by OsHKT2;1 and SOS1. HKT1, ZmHAK4, and OsHAK12 transport Na+ from xylem vessels to parenchyma cells. SKOR mediates K+ loading into the xylem; while ZmHKT2 removes K+ from the xylem. NPF2.4 and SLAH1/3 mediate the loading and unloading of Cl- in the root xylem

图4 植物调节质膜H+-ATP酶活性响应碱性盐胁迫的分子机制 小肽(Peps)与其受体(PEPRs)感知胞外pH变化,激活下游信号通路。碱性盐胁迫下,胞质中Ca2+浓度升高,SCaBP3感知Ca2+信号,解除SCaBP3对AHA2的抑制,此外J3可以抑制PKS5对AHA2的抑制。Ca2+与ZmNSA1结合,诱导其通过26S蛋白酶体途径降解,使MHA2和MHA4表达水平上调。MPK级联途径调节盐碱胁迫下PM H+-ATP酶的转录水平和蛋白活性。碱胁迫下TaCCD1和TaSAUR215相互作用,抑制TaPP2C.D1/8对TaHA2的去磷酸化,增强TaHA2的活性
Fig. 4 Molecular mechanism of regulating plasma membrane H+-ATPase activity in plant response to alkaline salt stress Small peptides(Peps)and receptors(PEPRs)sense changes in extracellular pH and activate downstream signaling pathways. Under alkaline salt stress, the cytosolic Ca2+ concentration increases, and SCaBP3 senses the Ca2+ signal and relieves the inhibition of AHA2. Moreover, J3 can inhibit the inhibition of AHA2 by PKS5. Ca2+ binds to ZmNSA1 and induces its degradation through the 26S proteasome pathway, upregulating the expression levels of MHA2 and MHA4. The MPK cascade pathway regulates the transcriptional level and protein activity of PM H+-ATPase under salt stress. Under alkali stress, TaCCD1 and TaSAUR215 interact to inhibit the dephosphorylation of TaPP2C.D1/8 on TaHA2 and enhance the activity of TaHA2
| [1] |
Ismail AM, Horie T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance[J]. Annu Rev Plant Biol, 2017, 68: 405-434.
doi: 10.1146/annurev-arplant-042916-040936 pmid: 28226230 |
| [2] | 云雪雪, 陈雨生. 国际盐碱地开发动态及其对我国的启示[J]. 国土与自然资源研究, 2020(1): 84-87. |
| Yun XX, Chen YS. International development of saline-alkali land and its enlightenment to China[J]. Territ Nat Resour Study, 2020(1): 84-87. | |
| [3] | Zörb C, Geilfus CM, Dietz KJ. Salinity and crop yield[J]. Plant Biol, 2018, 21: 31-38. |
| [4] | 马国辉, 郑殿峰, 母德伟, 等. 耐盐碱水稻研究进展与展望[J]. 杂交水稻, 2024, 39(1): 1-10. |
| Ma GH, Zheng DF, Mu DW, et al. Research progress and prospect of saline-alkali tolerant rice[J]. Hybrid Rice, 2024, 39(1): 1-10. | |
| [5] | 王佺珍, 刘倩, 高娅妮, 等. 植物对盐碱胁迫的响应机制研究进展[J]. 生态学报, 2017, 37(16): 5565-5577. |
| Wang QZ, Liu Q, Gao YN, et al. Review on the mechanisms of the response to salinity-alkalinity stress in plants[J]. Acta Ecol Sin, 2017, 37(16): 5565-5577. | |
| [6] | Guo R, Yang ZZ, Li F, et al. Comparative metabolic responses and adaptive strategies of wheat(Triticum aestivum)to salt and alkali stress[J]. BMC Plant Biol, 2015, 15: 170. |
| [7] | Shi DC, Wang DL. Effects of various salt-alkaline mixed stresses on Aneurolepidium chinense(Trin.) Kitag[J]. Plant Soil, 2005, 271(1): 15-26. |
| [8] | Li H, Shi JY, Wang ZP, et al. H2S pretreatment mitigates the alkaline salt stress on Malus hupehensis roots by regulating Na+/K+ homeostasis and oxidative stress[J]. Plant Physiol Biochem, 2020, 156: 233-241. |
| [9] | Li XY, Li SX, Wang JH, et al. Exogenous abscisic acid alleviates harmful effect of salt and alkali stresses on wheat seedlings[J]. Int J Environ Res Public Health, 2020, 17(11): 3770. |
| [10] | Zhang S, Wu QR, Liu LL, et al. Osmotic stress alters circadian cytosolic Ca2+ oscillations and OSCA1 is required in circadian gated stress adaptation[J]. Plant Signal Behav, 2020, 15(12): 1836883. |
| [11] | Jiang ZH, Zhou XP, Tao M, et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx[J]. Nature, 2019, 572(7769): 341-346. |
| [12] | Chen K, Gao JH, Sun SJ, et al. BONZAI proteins control global osmotic stress responses in plants[J]. Curr Biol, 2020, 30(24): 4815-4825.e4. |
| [13] | Yan JW, Liu Y, Yan JW, et al. The salt-activated CBF1/CBF2/CBF3-GALS1 module fine-tunes galactan-induced salt hypersensitivity in Arabidopsis[J]. J Integr Plant Biol, 2023, 65(8): 1904-1917. |
| [14] | Yan JW, Liu Y, Yang L, et al. Cell wall β-1, 4-galactan regulated by the BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana[J]. Mol Plant, 2021, 14(3): 411-425. |
| [15] | Hsu PK, Dubeaux G, Takahashi Y, et al. Signaling mechanisms in abscisic acid-mediated stomatal closure[J]. Plant J, 2021, 105(2): 307-321. |
| [16] |
Yoshida T, Mogami J, Yamaguchi-Shinozaki K. Omics approaches toward defining the comprehensive abscisic acid signaling network in plants[J]. Plant Cell Physiol, 2015, 56(6): 1043-1052.
doi: 10.1093/pcp/pcv060 pmid: 25917608 |
| [17] |
Sah SK, Reddy KR, Li JX. Abscisic acid and abiotic stress tolerance in crop plants[J]. Front Plant Sci, 2016, 7: 571.
doi: 10.3389/fpls.2016.00571 pmid: 27200044 |
| [18] | Wang YH, Que F, Li T, et al. DcABF3, an ABF transcription factor from carrot, alters stomatal density and reduces ABA sensitivity in transgenic Arabidopsis[J]. Plant Sci, 2021, 302: 110699. |
| [19] | Jing YN, Yu Y, Wang HZ, et al. Genome-wide identification and expression analysis of the bZIP gene family in silver birch(Betula pendula Roth.)[J]. J For Res, 2022, 33(5): 1615-1636. |
| [20] | Pan XJ, Wang CL, Liu ZS, et al. Identification of ABF/AREB gene family in tomato(Solanum lycopersicum L.) and functional analysis of ABF/AREB in response to ABA and abiotic stresses[J]. PeerJ, 2023, 11: e15310. |
| [21] | Jin DJ, Li SQ, Li ZP, et al. Arabidopsis ABRE-binding factors modulate salinity-induced inhibition of root hair growth by interacting with and suppressing RHD6[J]. Plant Sci, 2023, 332: 111728. |
| [22] | Pan WC, Zheng PP, Zhang C, et al. The effect of abre binding factor 4-mediated fyve1 on salt stress tolerance in Arabidopsis[J]. Plant Sci, 2020, 296: 110489. |
| [23] |
Dong T, Park Y, Hwang I. Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling[J]. Essays Biochem, 2015, 58: 29-48.
doi: 10.1042/bse0580029 pmid: 26374885 |
| [24] |
Chen XX, Ding YL, Yang YQ, et al. Protein kinases in plant responses to drought, salt, and cold stress[J]. J Integr Plant Biol, 2021, 63(1): 53-78.
doi: 10.1111/jipb.13061 |
| [25] |
Niu ML, Sun ST, Nawaz MA, et al. Grafting cucumber onto pumpkin induced early stomatal closure by increasing ABA sensitivity under salinity conditions[J]. Front Plant Sci, 2019, 10: 1290.
doi: 10.3389/fpls.2019.01290 pmid: 31781131 |
| [26] |
Wang PC, Zhao Y, Li ZP, et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response[J]. Mol Cell, 2018, 69(1): 100-112.e6.
doi: S1097-2765(17)30930-9 pmid: 29290610 |
| [27] | Su Y, Luo WG, Lin WH, et al. Model of cation transportation mediated by high-affinity potassium transporters(HKTs)in higher plants[J]. Biol Proced Online, 2015, 17: 1. |
| [28] |
Munns R, Passioura JB, Colmer TD, et al. Osmotic adjustment and energy limitations to plant growth in saline soil[J]. New Phytol, 2020, 225(3): 1091-1096.
doi: 10.1111/nph.15862 pmid: 31006123 |
| [29] |
Batelli G, Verslues PE, Agius F, et al. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity[J]. Mol Cell Biol, 2007, 27(22): 7781-7790.
doi: 10.1128/MCB.00430-07 pmid: 17875927 |
| [30] | Cao YB, Zhang M, Liang XY, et al. Natural variation of an EF-hand Ca2+-binding-protein coding gene confers saline-alkaline tolerance in maize[J]. Nat Commun, 2020, 11(1): 186. |
| [31] | Kumar G, Basu S, Singla-Pareek SL, et al. Unraveling the contribution of OsSOS2 in conferring salinity and drought tolerance in a high-yielding rice[J]. Physiol Plant, 2022, 174(1): e13638. |
| [32] | Quan RD, Lin HX, Mendoza I, et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress[J]. Plant Cell, 2007, 19(4): 1415-1431. |
| [33] |
Steinhorst L, He GF, Moore LK, et al. A Ca2+-sensor switch for tolerance to elevated salt stress in Arabidopsis[J]. Dev Cell, 2022, 57(17): 2081-2094.e7.
doi: 10.1016/j.devcel.2022.08.001 pmid: 36007523 |
| [34] | Yin XC, Xia YQ, Xie Q, et al. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance[J]. J Exp Bot, 2020, 71(6): 1801-1814. |
| [35] | Xie Q, Yin XC, Wang Y, et al. The signalling pathways, calcineurin B-like protein 5(CBL5)-CBL-interacting protein kinase 8(CIPK8)/CIPK24-salt overly sensitive 1(SOS1), transduce salt signals in seed germination in Arabidopsis[J]. Plant Cell Environ, 2024, 47(5): 1486-1502. |
| [36] | Zhou HP, Lin HX, Chen S, et al. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins[J]. Plant Cell, 2014, 26(3): 1166-1182. |
| [37] |
Yang ZJ, Wang CW, Xue Y, et al. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance[J]. Nat Commun, 2019, 10(1): 1199.
doi: 10.1038/s41467-019-09181-2 pmid: 30867421 |
| [38] | Cha JY, Kim J, Jeong SY, et al. The Na+/H+ antiporter salt overly sensitive 1 regulates salt compensation of circadian rhythms by stabilizing GIGANTEA in Arabidopsis[J]. Proc Natl Acad Sci U S A, 2022, 119(33): e2207275119. |
| [39] |
Ma L, Ye JM, Yang YQ, et al. The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress[J]. Dev Cell, 2019, 48(5): 697-709.e5.
doi: S1534-5807(19)30101-7 pmid: 30861376 |
| [40] |
Li JF, Zhou XY, Wang Y, et al. Inhibition of the maize salt overly sensitive pathway by ZmSK3 and ZmSK4[J]. J Genet Genomics, 2023, 50(12): 960-970.
doi: 10.1016/j.jgg.2023.04.010 pmid: 37127254 |
| [41] | Plasencia FA, Estrada Y, Flores FB, et al. The Ca2+ sensor calcineurin B-like protein 10 in plants: emerging new crucial roles for plant abiotic stress tolerance[J]. Front Plant Sci, 2021, 11: 599944. |
| [42] | Barragán V, Leidi EO, Andrés Z, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis[J]. Plant Cell, 2012, 24(3): 1127-1142. |
| [43] | Sun MH, Ma QJ, Hu DG, et al. The glucose sensor MdHXK1 phosphorylates a tonoplast Na+/H+ exchanger to improve salt tolerance[J]. Plant Physiol, 2018, 176(4): 2977-2990. |
| [44] | Ding CJ, Zhang WX, Li D, et al. Effect of overexpression of JERFs on intracellular K+/Na+ balance in transgenic poplar(Populus alba × P. berolinensis)under salt stress[J]. Front Plant Sci, 2020, 11: 1192. |
| [45] |
Sunarpi, Horie T, Motoda J, et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells[J]. Plant J, 2005, 44(6): 928-938.
doi: 10.1111/j.1365-313X.2005.02595.x pmid: 16359386 |
| [46] | Byrt CS, Xu B, Krishnan M, et al. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat[J]. Plant J, 2014, 80(3): 516-526. |
| [47] | Ren ZJ, Liu Y, Kang D, et al. Two alternative splicing variants of maize HKT1;1 confer salt tolerance in transgenic tobacco plants[J]. Plant Cell Tissue Organ Cult PCTOC, 2015, 123(3): 569-578. |
| [48] | Kobayashi NI, Yamaji N, Yamamoto H, et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice[J]. Plant J, 2017, 91(4): 657-670. |
| [49] |
Henderson SW, Dunlevy JD, Wu Y, et al. Functional differences in transport properties of natural HKT1;1 variants influence shoot Na+ exclusion in grapevine rootstocks[J]. New Phytol, 2018, 217(3): 1113-1127.
doi: 10.1111/nph.14888 pmid: 29160564 |
| [50] |
Cao YB, Zhou XY, Song HF, et al. Advances in deciphering salt tolerance mechanism in maize[J]. Crop J, 2023, 11(4): 1001-1010..
doi: 10.1016/j.cj.2022.12.004 |
| [51] | Song HF, Cao YB, Zhao XY, et al. Na+-preferential ion transporter HKT1;1 mediates salt tolerance in blueberry[J]. Plant Physiol, 2023, 194(1): 511-529. |
| [52] | Baek D, Jiang JF, Chung JS, et al. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance[J]. Plant Cell Physiol, 2011, 52(1): 149-161. |
| [53] | Wang J, Nan N, Li N, et al. A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress[J]. Plant Cell, 2020, 32(11): 3535-3558. |
| [54] |
Liu YT, Chen X, Xue SY, et al. SET DOMAIN GROUP 721 protein functions in saline-alkaline stress tolerance in the model rice variety Kitaake[J]. Plant Biotechnol J, 2021, 19(12): 2576-2588.
doi: 10.1111/pbi.13683 pmid: 34416090 |
| [55] |
Yu J, Zhu CS, Xuan W, et al. Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice[J]. Nat Commun, 2023, 14(1): 3550.
doi: 10.1038/s41467-023-39167-0 pmid: 37321989 |
| [56] | Chu ML, Chen PW, Meng SF, et al. The Arabidopsis phosphatase PP2C49 negatively regulates salt tolerance through inhibition of AtHKT1;1[J]. J Integr Plant Biol, 2021, 63(3): 528-542. |
| [57] | Liu YT, Li MT, Yu JL, et al. Plasma membrane-localized Hsp40/DNAJ chaperone protein facilitates OsSUVH7-OsBAG4-OsMYB106 transcriptional complex formation for OsHKT1;5 activation[J]. J Integr Plant Biol, 2023, 65(1): 265-279. |
| [58] | Horie T, Costa A, Kim TH, et al. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth[J]. EMBO J, 2007, 26(12): 3003-3014. |
| [59] | Wang XH, Shen XS, Qu YN, et al. Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters[J]. Nat Plants, 2024, 10(4): 633-644. |
| [60] |
Zhang M, Liang XY, Wang LM, et al. A HAK family Na+ transporter confers natural variation of salt tolerance in maize[J]. Nat Plants, 2019, 5(12): 1297-1308.
doi: 10.1038/s41477-019-0565-y pmid: 31819228 |
| [61] | Zhang LN, Sun XY, Li YF, et al. Rice Na+-permeable transporter OsHAK12 mediates shoots Na+ exclusion in response to salt stress[J]. Front Plant Sci, 2021, 12: 771746. |
| [62] | Wang Z, Hong YC, Zhu GT, et al. Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter[J]. EMBO J, 2020, 39(10): e103256. |
| [63] | Uddin N, Li X, Ullah MW, et al. Lignin developmental patterns and Casparian strip as apoplastic barriers: a review[J]. Int J Biol Macromol, 2024, 260(Pt 2): 129595. |
| [64] | Chen AL, Liu T, Wang Z, et al. Plant root suberin: a layer of defence against biotic and abiotic stresses[J]. Front Plant Sci, 2022, 13: 1056008. |
| [65] | Chen CX, He GF, Li JF, et al. A salt stress-activated GSO1-SOS2-SOS1 module protects the Arabidopsis root stem cell niche by enhancing sodium ion extrusion[J]. EMBO J, 2023, 42(13): e113004. |
| [66] |
Wang YY, Cao YB, Liang XY, et al. A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize[J]. Nat Commun, 2022, 13(1): 2222.
doi: 10.1038/s41467-022-29809-0 pmid: 35468878 |
| [67] | Gao J, Mol S, et al. The Arabidopsis GORK K+-channel is phosphorylated by calcium-dependent protein kinase 21(CPK21), which in turn is activated by 14-3-3 proteins[J]. Plant Physiol Biochem, 2018, 125: 219-231. |
| [68] |
Lefoulon C, Boeglin M, Moreau B, et al. The Arabidopsis AtPP2CA protein phosphatase inhibits the GORK K+ efflux channel and exerts a dominant suppressive effect on phosphomimetic-activating mutations[J]. J Biol Chem, 2016, 291(12): 6521-6533.
doi: 10.1074/jbc.M115.711309 pmid: 26801610 |
| [69] | Wu HH, Shabala L, Zhou MX, et al. Chloroplast-generated ROS dominate NaCl(-)induced K(+)efflux in wheat leaf mesophyll[J]. Plant Signal Behav, 2015, 10(5): e1013793. |
| [70] |
Horie T, Sugawara M, Okada T, et al. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells[J]. J Biosci Bioeng, 2011, 111(3): 346-356.
doi: 10.1016/j.jbiosc.2010.10.014 pmid: 21084222 |
| [71] | Chen G, Hu QD, Luo L, et al. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges[J]. Plant Cell Environ, 2015, 38(12): 2747-2765. |
| [72] | Shen Y, Shen LK, Shen ZX, et al. The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice[J]. Plant Cell Environ, 2015, 38(12): 2766-2779. |
| [73] | Okada T, Yamane S, Yamaguchi M, et al. Characterization of rice KT/HAK/KUP potassium transporters and K+ uptake by HAK1 from Oryza sativa[J]. Plant Biotechnol, 2018, 35(2): 101-111. |
| [74] | Ren XL, Qi GN, Feng HQ, et al. Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis[J]. Plant J, 2013, 74(2): 258-266. |
| [75] | Li JF, Shen LK, Han XL, et al. Phosphatidic acid-regulated SOS2 controls sodium and potassium homeostasis in Arabidopsis under salt stress[J]. EMBO J, 2023, 42(8): e112401. |
| [76] |
Nieves-Cordones M, Amo J, Hurtado-Navarro L, et al. Inhibition of SlSKOR by SlCIPK23-SlCBL1/9 uncovers CIPK-CBL-target network rewiring in land plants[J]. New Phytol, 2023, 238(6): 2495-2511.
doi: 10.1111/nph.18910 pmid: 36967582 |
| [77] | Brodsky DE. SLAC1-related signal transduction pathway involved in ABA- induced stomatal closure and K+ selective transport by the OsHKT2;4 transporter from rice(Oryza sativa)with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions[D]. California: UC San Diego, 2011. |
| [78] |
Sassi A, Mieulet D, Khan I, et al. The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity[J]. Plant Physiol, 2012, 160(1): 498-510.
doi: 10.1104/pp.112.194936 pmid: 22773759 |
| [79] | Cao YB, Liang XY, Yin P, et al. A domestication-associated reduction in K+-preferring HKT transporter activity underlies maize shoot K+ accumulation and salt tolerance[J]. New Phytol, 2019, 222(1): 301-317. |
| [80] | Geilfus CM. Chloride: from nutrient to toxicant[J]. Plant Cell Physiol, 2018, 59(5): 877-886. |
| [81] |
Ren ZJ, Bai FL, Xu JW, et al. A chloride efflux transporter, BIG RICE GRAIN 1, is involved in mediating grain size and salt tolerance in rice[J]. J Integr Plant Biol, 2021, 63(12): 2150-2163.
doi: 10.1111/jipb.13178 |
| [82] |
Franco-Navarro JD, Brumós J, Rosales MA, et al. Chloride regulates leaf cell size and water relations in tobacco plants[J]. J Exp Bot, 2016, 67(3): 873-891.
doi: 10.1093/jxb/erv502 pmid: 26602947 |
| [83] | Wu HH, Li ZH. The importance of Cl- exclusion and vacuolar Cl- sequestration: revisiting the role of Cl- transport in plant salt tolerance[J]. Front Plant Sci, 2019, 10: 1418. |
| [84] | Li B, Byrt C, Qiu JE, et al. Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis[J]. Plant Physiol, 2016, 170(2): 1014-1029. |
| [85] | Li B, Qiu JE, Jayakannan M, et al. AtNPF2.5 modulates chloride(Cl-)efflux from roots of Arabidopsis thaliana[J]. Front Plant Sci, 2017, 7: 2013. |
| [86] |
Cubero-Font P, Maierhofer T, Jaslan J, et al. Silent S-type anion channel subunit SLAH1 gates SLAH3 open for chloride root-to-shoot translocation[J]. Curr Biol, 2016, 26(16): 2213-2220.
doi: 10.1016/j.cub.2016.06.045 pmid: 27397895 |
| [87] | Qiu JE, Henderson SW, Tester M, et al. SLAH1, a homologue of the slow type anion channel SLAC1, modulates shoot Cl- accumulation and salt tolerance in Arabidopsis thaliana[J]. J Exp Bot, 2016, 67(15): 4495-4505. |
| [88] | Colmenero-Flores JM, Franco-Navarro JD, Cubero-Font P, et al. Chloride as a beneficial macronutrient in higher plants: new roles and regulation[J]. Int J Mol Sci, 2019, 20(19): 4686. |
| [89] |
Henderson SW, Wege S, Qiu JE, et al. Grapevine and Arabidopsis cation-chloride cotransporters localize to the Golgi and trans-golgi network and indirectly influence long-distance ion transport and plant salt tolerance[J]. Plant Physiol, 2015, 169(3): 2215-2229.
doi: 10.1104/pp.15.00499 pmid: 26378102 |
| [90] | McKay DW, McFarlane HE, Qu Y, et al. Plant Trans-Golgi Network/Early Endosome pH regulation requires Cation Chloride Cotransporter(CCC1)[J]. eLife, 2022, 11: e70701. |
| [91] | Jossier M, Kroniewicz L, Dalmas F, et al. The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance[J]. Plant J, 2010, 64(4): 563-576. |
| [92] |
Nguyen CT, Agorio A, Jossier M, et al. Characterization of the chloride channel-like, AtCLCg, involved in chloride tolerance in Arabidopsis thaliana[J]. Plant Cell Physiol, 2016, 57(4): 764-775.
doi: 10.1093/pcp/pcv169 pmid: 26556649 |
| [93] | De Angeli A, Zhang JB, Meyer S, et al. AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis[J]. Nat Commun, 2013, 4: 1804. |
| [94] | Yin P, Liang XY, Zhao HS, et al. Cytokinin signaling promotes salt tolerance by modulating shoot chloride exclusion in maize[J]. Mol Plant, 2023, 16(6): 1031-1047. |
| [95] | Mimata Y, Munemasa S, Akter F, et al. Malate induces stomatal closure via a receptor-like kinase GHR1- and reactive oxygen species-dependent pathway in Arabidopsis thaliana[J]. Biosci Biotechnol Biochem, 2022, 86(10): 1362-1367. |
| [96] | Liu P, Wu XL, Gong BB, et al. Review of the mechanisms by which transcription factors and exogenous substances regulate ROS metabolism under abiotic stress[J]. Antioxidants, 2022, 11(11): 2106. |
| [97] | Wang LQ, Zhang CR, Wang YM, et al. Tamarix hispida aquaporin ThPIP2;5 confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis[J]. Environ Exp Bot, 2018, 152: 158-166. |
| [98] | Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants[J]. Plant Physiol Biochem, 2010, 48(12): 909-930. |
| [99] | Hasanuzzaman M, Bhuyan MHMB, Anee TI, et al. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress[J]. Antioxidants, 2019, 8(9): 384. |
| [100] | Kohli SK, Khanna K, Bhardwaj R, et al. Assessment of subcellular ROS and NO metabolism in higher plants: multifunctional signaling molecules[J]. Antioxidants, 2019, 8(12): 641. |
| [101] | Suo JW, Zhang H, Zhao Q, et al. Na2CO3-responsive photosynthetic and ROS scavenging mechanisms in chloroplasts of alkaligrass revealed by phosphoproteomics[J]. Genomics Proteomics Bioinformatics, 2020, 18(3): 271-288. |
| [102] | Sharma A, Shahzad B, Kumar V, et al. Phytohormones regulate accumulation of osmolytes under abiotic stress[J]. Biomolecules, 2019, 9(7): 285. |
| [103] | Hasanuzzaman M, Alam MM, Rahman A, et al. Exogenous proline and glycine betaine mediated upregulation of antioxidant defense and glyoxalase systems provides better protection against salt-induced oxidative stress in two rice(Oryza sativa L.) varieties[J]. Biomed Res Int, 2014, 2014: 757219. |
| [104] | Zhao K, Cheng ZH, Guo Q, et al. Characterization of the poplar R2R3-MYB gene family and over-expression of PsnMYB108 confers salt tolerance in transgenic tobacco[J]. Front Plant Sci, 2020, 11: 571881. |
| [105] | Liu L, Song W, Huang SJ, et al. Extracellular pH sensing by plant cell-surface peptide-receptor complexes[J]. Cell, 2022, 185(18): 3341-3355.e13. |
| [106] | Zhang HL, Yu FF, Xie P, et al. A Gγ protein regulates alkaline sensitivity in crops[J]. Science, 2023, 379(6638): eade8416. |
| [107] | Duan XB, Yu Y, Zhang Y, et al. A potential efflux boron transporter gene GsBOR2, positively regulates Arabidopsis bicarbonate tolerance[J]. Plant Sci, 2018, 274: 284-292. |
| [108] | Cao L, Yu Y, DuanMu HZ, et al. A novel Glycine soja homeodomain-leucine zipper(HD-Zip)I gene, Gshdz4, positively regulates bicarbonate tolerance and responds to osmotic stress in Arabidopsis[J]. BMC Plant Biol, 2016, 16(1): 184. |
| [109] | Liu D, Ma Y, Rui MM, et al. Is high pH the key factor of alkali stress on plant growth and physiology? A case study with wheat(Triticum aestivum L.) seedlings[J]. Agronomy, 2022, 12(8): 1820. |
| [110] | Chen C, Sun XL, Duanmu HZ, et al. GsCML27, a gene encoding a calcium-binding ef-hand protein from Glycine soja, plays differential roles in plant responses to bicarbonate, salt and osmotic stresses[J]. PLoS One, 2015, 10(11): e0141888. |
| [111] | Tian W, Hou CC, Ren ZJ, et al. A molecular pathway for CO2 response in Arabidopsis guard cells[J]. Nat Commun, 2015, 6: 6057. |
| [112] | Janicka M, Wdowikowska A, Kłobus G. Assay of plasma membrane H+-ATPase in plant tissues under abiotic stresses[J]. Methods Mol Biol, 2018, 1696: 205-215. |
| [113] | 刘奕媺, 于洋, 方军. 盐碱胁迫及植物耐盐碱分子机制研究[J]. 土壤与作物, 2018, 7(2): 201-211. |
| Liu YM, Yu Y, Fang J. Saline-alkali stress and molecular mechanism of saline-alkali tolerance in plants[J]. Soils Crops, 2018, 7(2): 201-211. | |
| [114] | Zhou LJ, Zhang CL, Zhang RF, et al. The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis[J]. Plant Physiol, 2019, 179(1): 88-106. |
| [115] | Kinoshita SN, Suzuki T, Kiba T, et al. Photosynthetic-product-dependent activation of plasma membrane H+-ATPase and nitrate uptake in Arabidopsis leaves[J]. Plant Cell Physiol, 2023, 64(2): 191-203. |
| [116] | Jia BW, Cui HL, Zhang DJ, et al. The conserved evolution of plant H+-ATPase family and the involvement of soybean H+-ATPases in sodium bicarbonate stress responses[J]. Plant Physiol Biochem, 2023, 204: 108133. |
| [117] |
Fuglsang AT, Guo Y, Cuin TA, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein[J]. Plant Cell, 2007, 19(5): 1617-1634.
doi: 10.1105/tpc.105.035626 pmid: 17483306 |
| [118] | Lin HX, Du WM, Yang YQ, et al. A calcium-independent activation of the Arabidopsis SOS2-like protein kinase24 by its interacting SOS3-like calcium binding protein1[J]. Plant Physiol, 2014, 164(4): 2197-2206. |
| [119] | Yang YQ, Qin YX, Xie CG, et al. The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase[J]. Plant Cell, 2010, 22(4): 1313-1332. |
| [120] | Pallucca R, Visconti S, Camoni L, et al. Specificity of ε and non-ε isoforms of Arabidopsis 14-3-3 proteins towards the H+-ATPase and other targets[J]. PLoS One, 2014, 9(6): e90764. |
| [121] | Yang YQ, Wu YJ, Ma L, et al. The Ca2+ sensor SCaBP3/CBL7 modulates plasma membrane H+-ATPase activity and promotes alkali tolerance in Arabidopsis[J]. Plant Cell, 2019, 31(6): 1367-1384. |
| [122] | Liu X, Wu YJ, Fu HQ, et al. SCaBP3/CBL7 negatively regulates the plasma membrane H+-ATPase and modulates hypocotyl elongation in Arabidopsis[J]. Plant Signal Behav, 2022, 17(1): 2092699. |
| [123] | Cui MH, Li YP, Li JH, et al. Ca2+-dependent TaCCD1 cooperates with TaSAUR215 to enhance plasma membrane H+-ATPase activity and alkali stress tolerance by inhibiting PP2C-mediated dephosphorylation of TaHA2 in wheat[J]. Mol Plant, 2023, 16(3): 571-587. |
| [124] | Sun QR, Zhao DR, Gao M, et al. MxMPK6-2-mediated phosphorylation enhances the responseof apple rootstocks to Fe deficiency by activating PM H+-ATPase MxHA2[J]. Plant J, 2023, 116(1): 69-86. |
| [125] |
Lin HX, Zhu MZ, Yano M, et al. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance[J]. Theor Appl Genet, 2004, 108(2): 253-260.
doi: 10.1007/s00122-003-1421-y pmid: 14513218 |
| [126] | He YQ, Yang B, He Y, et al. A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice[J]. Plant J, 2019, 97(6): 1089-1104. |
| [127] |
Zhang M, Cao YB, Wang ZP, et al. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize[J]. New Phytol, 2018, 217(3): 1161-1176.
doi: 10.1111/nph.14882 pmid: 29139111 |
| [128] | Zhang M, Li YD, Liang XY, et al. A teosinte-derived allele of an HKT1 family sodium transporter improves salt tolerance in maize[J]. Plant Biotechnol J, 2023, 21(1): 97-108. |
| [129] | Huang SB, Spielmeyer W, Lagudah ES, et al. A sodium transporter(HKT7)is a candidate for Nax1, a gene for salt tolerance in durum wheat[J]. Plant Physiol, 2006, 142(4): 1718-1727. |
| [130] | James RA, Blake C, Byrt CS, et al. Major genes for Na+ exclusion, Nax1 and Nax2(wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions[J]. J Exp Bot, 2011, 62(8): 2939-2947. |
| [131] |
Munns R, James RA, Xu B, et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene[J]. Nat Biotechnol, 2012, 30(4): 360-364.
doi: 10.1038/nbt.2120 pmid: 22407351 |
| [132] | Wang M, Cheng J, Wu JH, et al. Variation in TaSPL6-D confers salinity tolerance in bread wheat by activating TaHKT1;5-D while preserving yield-related traits[J]. Nat Genet, 2024, 56(6): 1257-1269. |
| [133] | Luo TT, Ma CX, Fan YH, et al. CRISPR-Cas9-mediated editing of GmARM improves resistance to multiple stresses in soybean[J]. Plant Sci, 2024, 346: 112147. |
| [134] | Mohamed HI, Khan A, Basit A. CRISPR-Cas9 system mediated genome editing technology: an ultimate tool to enhance abiotic stress in crop plants[J]. J Soil Sci Plant Nutr, 2024, 24(2): 1799-1822. |
| [135] | Yang MZ, Zhou BY, Song ZG, et al. A calmodulin-like protein PvCML9 negatively regulates salt tolerance[J]. Plant Physiol Biochem, 2024, 210: 108642. |
| [136] | Ly LK, Ho TM, Bui TP, et al. CRISPR/Cas9 targeted mutations of OsDSG1 gene enhanced salt tolerance in rice[J]. Funct Integr Genomics, 2024, 24(2): 70. |
| [1] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [2] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [3] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [4] | 徐红云, 张明意. GRAS转录因子AtSCL4负调控拟南芥应答渗透胁迫[J]. 生物技术通报, 2022, 38(6): 129-135. |
| [5] | 胡华冉, 杜磊, 张芮豪, 钟秋月, 刘发万, 桂敏. 辣椒适应非生物胁迫的研究进展[J]. 生物技术通报, 2022, 38(12): 58-72. |
| [6] | 姜焕焕, 王通, 陈娜, 禹山林, 迟晓元, 王冕, 祁佩时. 根际促生菌提高植物抗盐碱性的研究进展[J]. 生物技术通报, 2019, 35(10): 189-197. |
| [7] | 王娜, 龚娜, 刘国丽, 马晓颖, 杨镇, 杨涛. 内生菌次生代谢产物提高玉米渗透胁迫抗性的表达谱分析[J]. 生物技术通报, 2016, 32(7): 87-92. |
| [8] | 刘丽萍, 张东智, 张冲, 陈金焕. 黑果枸杞抗逆性及栽培育种研究进展[J]. 生物技术通报, 2016, 32(10): 118-127. |
| [9] | 李娇, 张宝龙, 赵颖, 辛士刚, 陈强, 宋宁宁, 李雪梅. 内生菌对提高植物抗盐碱性的研究进展[J]. 生物技术通报, 2014, 30(4): 14-18. |
| [10] | 胡海英;石晶;戴海霞;蔡倩;崔雪琼;冯秀娟;黄琦;姚新灵;. E.coli中表达马铃薯OSM-3b基因对增强渗透胁迫抗性的分析[J]. , 2012, 0(06): 66-70. |
| [11] | 何宝坤;王玉洁;孙立平. 蛋白质组学研究技术及其在植物抗渗透胁迫研究中的应用[J]. , 2005, 0(02): 18-21. |
| [12] | 何宝坤;王玉洁;孙立平. 蛋白质组学研究技术及其在植物抗渗透胁迫研究中的应用[J]. , 2005, 0(02): 18-21. |
| [13] | . 食品上的应用[J]. , 1995, 0(04): 98-107. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||