








生物技术通报 ›› 2024, Vol. 40 ›› Issue (11): 227-235.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0318
收稿日期:2024-03-31
出版日期:2024-11-26
发布日期:2024-12-19
通讯作者:
熊王丹,女,博士,研究方向:植物分子育种;E-mail: xiongwd@qau.edu.cn作者简介:王其昕,男,研究方向:植物分子育种;E-mail: 2730926732@qq.com
基金资助:
WANG Qi-xin1( ), ZHANG Ling-xin1, ZHONG Ni-na1, XIONG Wang-dan1,2(
), ZHANG Ling-xin1, ZHONG Ni-na1, XIONG Wang-dan1,2( )
)
Received:2024-03-31
Published:2024-11-26
Online:2024-12-19
摘要:
【目的】 生长素响应因子ARF(auxin response factor)可以调控生长素响应基因的表达,在植物生长发育中发挥着重要作用。从狗尾草全基因组范围内鉴定出SvARF基因家族成员,分析相关基因表达特征,为后续深入研究其与狗尾草生长发育的关系提供支撑。【方法】 利用生物信息学方法从狗尾草基因组中鉴定了ARF转录因子成员,分析其系统进化、染色体定位和基因结构等情况。利用RT-qPCR(quantitative real-time polymerase chain reaction)分析该家族成员在生长素类除草剂2,4-D异辛酯处理后的表达响应情况。【结果】 共鉴定到24个狗尾草SvARF基因家族成员,可分成4个亚家族,同一亚家族SvARF基因的结构相似且基因表达模式相近。不同SvARF基因的表达模式存在差异,且有组织特异性,III亚家族的SvARF基因在根茎分蘖处的表达量均较高。2,4-D异辛酯处理后,有9个SvARF基因的相对表达量发生显著变化,其中SvARF5、SvARF6、SvARF9、SvARF10、SvARF17和SvARF18表达量显著上调,SvARF14、SvARF22和SvARF24表达量显著下调。【结论】 狗尾草SvARF基因家族有24个成员。生长素类除草剂影响SvARF基因的表达,暗示SvARF基因在抑制狗尾草植株分蘖中发挥重要作用。
王其昕, 张凌馨, 钟妮娜, 熊王丹. 狗尾草ARF基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(11): 227-235.
WANG Qi-xin, ZHANG Ling-xin, ZHONG Ni-na, XIONG Wang-dan. Identification and Expression Analysis of ARF Gene Family in Setaria viridis[J]. Biotechnology Bulletin, 2024, 40(11): 227-235.
| 基因名称Gene name | 正向引物 Forword primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| SvARF5 | GCAGTTCCGTCACATCTACCG | CGAGGAGTCATCCCAGTCAA |
| SvARF6 | ACCCGTGGGAGTCATTTGTG | GATAGGTAGCGTGGATCGTTTC |
| SvARF9 | AATTGGCTCGTTCGATTACG | ACTTGCCCTTGGCTGCTGT |
| SvARF10 | TATTTGGGTTTCCTCTTGGTAG | TTCTGAACATTCTGGGTAGCCT |
| SvARF14 | CCCTTCTTGCTCAACATCACC | CTTCGGCAGTTCCTTAGTCG |
| SvARF17 | TTCTCCAAAGCCTGAGCAAT | CAAATCTTGGAACCCCGAAG |
| SvARF18 | TGATGGTGAACTAAGACTGGGTG | AGACTGGACGGTTGAGGCTC |
| SvARF22 | CCTGACCTCTGGCACCCTGAT | TCCCAAGCACAAGCACCCAC |
| SvARF24 | GGTTTCTCATCTCAAGGTCGTG | AGCCTCCGTGCTACTTTCAT |
| SvActin | CAGTGGACGCACAACAGGTAT | AGCAAGGTCAAGACGGAGAAT |
表1 荧光定量 PCR 引物
Table 1 Primer sequences for quantitative real time PCR
| 基因名称Gene name | 正向引物 Forword primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| SvARF5 | GCAGTTCCGTCACATCTACCG | CGAGGAGTCATCCCAGTCAA |
| SvARF6 | ACCCGTGGGAGTCATTTGTG | GATAGGTAGCGTGGATCGTTTC |
| SvARF9 | AATTGGCTCGTTCGATTACG | ACTTGCCCTTGGCTGCTGT |
| SvARF10 | TATTTGGGTTTCCTCTTGGTAG | TTCTGAACATTCTGGGTAGCCT |
| SvARF14 | CCCTTCTTGCTCAACATCACC | CTTCGGCAGTTCCTTAGTCG |
| SvARF17 | TTCTCCAAAGCCTGAGCAAT | CAAATCTTGGAACCCCGAAG |
| SvARF18 | TGATGGTGAACTAAGACTGGGTG | AGACTGGACGGTTGAGGCTC |
| SvARF22 | CCTGACCTCTGGCACCCTGAT | TCCCAAGCACAAGCACCCAC |
| SvARF24 | GGTTTCTCATCTCAAGGTCGTG | AGCCTCCGTGCTACTTTCAT |
| SvActin | CAGTGGACGCACAACAGGTAT | AGCAAGGTCAAGACGGAGAAT |
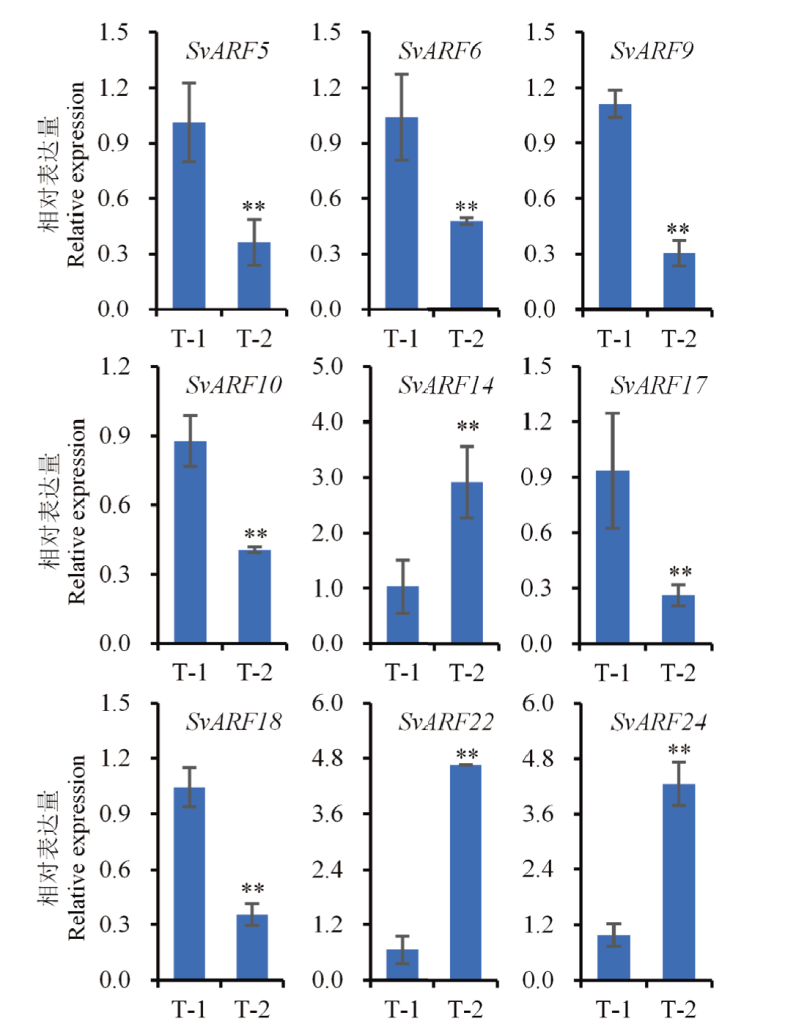
图7 喷施2,4-D异辛酯后SvARF基因在狗尾草分蘖中的相对表达情况 以SvARF基因在对照T-0时的表达量作为1,相应地计算处理时该基因表达水平。T-1和T-2分别表示2,4-D异辛酯处理浓度为1 050和4 200 g a.i./ha。**表示P<0.01,下同
Fig. 7 Relative expression analysis of SvARF genes in the tillers of S. viridis after spraying 2,4-D isooctyl ester The expression of SvARF genes under control T0-1 was defined as 1, and the relative expression of SvARF gene under treatment was determined by the relative fold changes between treatment and control. T-1 and T-2 refer to the concentration of 2,4-D isooctyl ester at 1 050 and 4 200 g a.i./ha, respectively. ** indicates P<0.01. The same below

图8 喷施2,4-D异辛酯后狗尾草分蘖中的IAA含量 T-0表示2,4-D异辛酯处理浓度为0 g a.i./ha
Fig. 8 Content of IAA in the tillers of S. viridis after spraying 2,4-D isooctyl ester T-0 refers to the concentration of 2,4-D isooctyl ester at 0 g a.i./ha
| [1] | Wang JH, Hu D, Shi XN, et al. Different responses of invasive weed Alternanthera philoxeroides and Oryza sativa to plant growth regulators[J]. Life, 2022, 12(7): 1069. |
| [2] |
Guo F, Huang YZ, Qi PP, et al. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance[J]. New Phytol, 2021, 229(5): 2676-2692.
doi: 10.1111/nph.17061 pmid: 33135782 |
| [3] |
Ishikawa S, Maekawa M, Arite T, et al. Suppression of tiller bud activity in tillering dwarf mutants of rice[J]. Plant Cell Physiol, 2005, 46(1): 79-86.
doi: 10.1093/pcp/pci022 pmid: 15659436 |
| [4] | Yu HY, Cui HL, Chen JC, et al. Regulation of 2, 4-D isooctyl ester on Triticum aestivum and Aegilops tauschii tillering and endogenous phytohormonal responses[J]. Front Plant Sci, 2021, 12: 642701. |
| [5] |
Roosjen M, Paque S, Weijers D. Auxin Response Factors: output control in auxin biology[J]. J Exp Bot, 2018, 69(2): 179-188.
doi: 10.1093/jxb/erx237 pmid: 28992135 |
| [6] |
Dinesh DC, Kovermann M, Gopalswamy M, et al. Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response[J]. Proc Natl Acad Sci USA, 2015, 112(19): 6230-6235.
doi: 10.1073/pnas.1424077112 pmid: 25918389 |
| [7] |
Li YH, Han S, Qi YH. Advances in structure and function of auxin response factor in plants[J]. J Integr Plant Biol, 2023, 65(3): 617-632.
doi: 10.1111/jipb.13392 |
| [8] |
Wilmoth JC, Wang SC, Tiwari SB, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation[J]. Plant J, 2005, 43(1): 118-130.
doi: 10.1111/j.1365-313X.2005.02432.x pmid: 15960621 |
| [9] |
Yang J, Tian L, Sun MX, et al. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis[J]. Plant Physiol, 2013, 162(2): 720-731.
doi: 10.1104/pp.113.214940 pmid: 23580594 |
| [10] | Ellis CM, Nagpal P, Young JC, et al. AUXIN RESPONSE FACTOR 1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana[J]. Development, 2005, 132(20): 4563-4574. |
| [11] | Sims K, Abedi-Samakush F, Szulc N, et al. OsARF11 promotes growth, meristem, seed, and vein formation during rice plant development[J]. Int J Mol Sci, 2021, 22(8): 4089. |
| [12] | Li PH, Brutnell TP. Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses[J]. J Exp Bot, 2011, 62(9): 3031-3037. |
| [13] |
Keshavarzi M, Seifali M. Systematic study of weedy species of Setaria(L.) P. Beauv.(Poaceae)in Iran[J]. Pak J Biol Sci, 2007, 10(19): 3362-3367.
pmid: 19090151 |
| [14] | Xiong WD, Xu XQ, Zhang L, et al. Genome-wide analysis of the WRKY gene family in physic nut(Jatropha curcas L.)[J]. Gene, 2013, 524(2): 124-132. |
| [15] | Zhao S, Zhang ML, Ma TL, et al. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress[J]. Plant Cell, 2016, 28(12): 3005-3019. |
| [16] | Chen Y, Liu B, Zhao YJ, et al. Whole-genome duplication and purifying selection contributes to the functional redundancy of auxin response factor(ARF)genes in foxtail millet(Setaria italica L.)[J]. Int J Genomics, 2021, 2021: 2590665. |
| [17] | Wang DK, Pei KM, Fu YP, et al. Genome-wide analysis of the auxin response factors(ARF)gene family in rice(Oryza sativa)[J]. Gene, 2007, 394(1/2): 13-24. |
| [18] | Song H, Guo ZL, Zhang XJ, et al. De novo genes in Arachis hypogaea cv. Tifrunner: systematic identification, molecular evolution, and potential contributions to cultivated peanut[J]. Plant J, 2022, 111(4): 1081-1095. |
| [19] | Liu R, Guo ZF, Lu SY. Genome-wide identification and expression analysis of the Aux/IAA and auxin response factor gene family in Medicago truncatula[J]. Int J Mol Sci, 2021, 22(19): 10494. |
| [20] | Chen FQ, Zhang JQ, Ha X, et al. Genome-wide identification and expression analysis of the auxin-response factor(ARF)gene family in Medicago sativa under abiotic stress[J]. BMC Genomics, 2023, 24(1): 498. |
| [21] |
Jiao YN. Double the genome, double the fun: genome duplications in angiosperms[J]. Mol Plant, 2018, 11(3): 357-358.
doi: S1674-2052(18)30059-5 pmid: 29476919 |
| [22] | Peng YK, Zhao K, Zheng RY, et al. A comprehensive analysis of auxin response factor gene family in Melastoma dodecandrum genome[J]. Int J Mol Sci, 2024, 25(2): 806. |
| [23] |
Okushima Y, Overvoorde PJ, Arima K, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19[J]. Plant Cell, 2005, 17(2): 444-463.
doi: 10.1105/tpc.104.028316 pmid: 15659631 |
| [24] | Wang HX, Ren J, Zhou SY, et al. Molecular regulation of oil gland development and biosynthesis of essential oils in Citrus spp[J]. Science, 2024, 383(6683): 659-666. |
| [25] |
Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm[J]. Annu Rev Cell Dev Biol, 2008, 24: 55-80.
doi: 10.1146/annurev.cellbio.23.090506.123214 pmid: 18631113 |
| [26] |
Korasick DA, Jez JM, Strader LC. Refining the nuclear auxin response pathway through structural biology[J]. Curr Opin Plant Biol, 2015, 27: 22-28.
doi: 10.1016/j.pbi.2015.05.007 pmid: 26048079 |
| [27] | Chandler JW. Auxin response factors[J]. Plant Cell Environ, 2016, 39(5): 1014-1028. |
| [28] |
Grossmann K. Auxin herbicides: current status of mechanism and mode of action[J]. Pest Manag Sci, 2010, 66(2): 113-120.
doi: 10.1002/ps.1860 pmid: 19823992 |
| [29] | Yan M, Yan Y, Wang P, et al. Genome-wide identification and expression analysis of auxin response factor(ARF)gene family in Panax ginseng indicates its possible roles in root development[J]. Plants, 2023, 12(23): 3943. |
| [30] | Li Y, Li JL, Chen ZH, et al. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice[J]. Plant Biotechnol J, 2020, 18(10): 2015-2026. |
| [31] | Li JC, Jiang YM, Zhang J, et al. Key auxin response factor(ARF)genes constraining wheat tillering of mutant dmc[J]. Peer J, 2021, 9: e12221. |
| [32] |
Su LY, Zhang T, Yang B, et al. Different evolutionary patterns of TIR1/AFBs and AUX/IAAs and their implications for the morphogenesis of land plants[J]. BMC Plant Biol, 2023, 23(1): 265.
doi: 10.1186/s12870-023-04253-4 pmid: 37202746 |
| [1] | 申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73. |
| [2] | 吴慧琴, 王延宏, 刘涵, 司政, 刘雪晴, 王静, 阳宜, 成妍. 辣椒UGT基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 198-211. |
| [3] | 满全财, 孟姿诺, 李伟, 蔡心汝, 苏润东, 付长青, 高顺娟, 崔江慧. 马铃薯AQP基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 51-63. |
| [4] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [5] | 周冉, 王兴平, 李彦霞, 罗仍卓么. 金黄色葡萄球菌型乳房炎奶牛乳腺组织的lncRNA差异表达分析[J]. 生物技术通报, 2024, 40(8): 320-328. |
| [6] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [7] | 崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82. |
| [8] | 刘丹丹, 王雷刚, 孙明慧, 焦小雨, 吴琼, 王文杰. 茶树海藻糖-6-磷酸合成酶(TPS)基因家族鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 152-163. |
| [9] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [10] | 李亦君, 杨小贝, 夏琳, 罗朝鹏, 徐馨, 杨军, 宁黔冀, 武明珠. 烟草NtPRR37基因克隆及功能分析[J]. 生物技术通报, 2024, 40(8): 221-231. |
| [11] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [12] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [13] | 任晓敏, 云岚, 艾芊, 赵乔. 新麦草异戊烯基转移酶PjIPT基因的功能验证[J]. 生物技术通报, 2024, 40(7): 207-215. |
| [14] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [15] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||