








生物技术通报 ›› 2025, Vol. 41 ›› Issue (10): 156-163.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0662
收稿日期:2025-06-23
出版日期:2025-10-26
发布日期:2025-10-28
通讯作者:
吴健,男,博士,副教授,研究方向 :油菜抗病分子遗传;E-mail: wu_jian@yzu.edu.cn作者简介:杨洋,女,研究方向 :植物学;E-mail: 2923977275@qq.com
基金资助:
YANG Yang( ), LIU Hui-min, LIN Li, WANG You-ping, WU Jian(
), LIU Hui-min, LIN Li, WANG You-ping, WU Jian( )
)
Received:2025-06-23
Published:2025-10-26
Online:2025-10-28
摘要:
目的 传统植物基因组DNA提取方法(如CTAB法、SDS裂解法等)存在操作繁琐、耗时长、使用有毒有机试剂等问题,难以满足当前高通量分子检测的实际需求。旨在开发一种快速、安全、适用于大规模样本处理的植物DNA提取方法,以提高分子检测效率。 方法 提出一种基于96深孔板的快速DNA提取方法——DDEB(directPCR DNA extraction buffer)法。该方法采用DDEB提取液,主要成分包括0.2 mol/L NaOH、0.01% SDS、50 mmol/L NaCl、0.1 mmol/L EDTA-2Na、0.15 g/L明胶及0.005%消泡剂A等组分。提取时将样品与提取液混合后通过振荡研磨,继而瞬时离心或静置沉降,即可获得可用于PCR扩增的DNA粗提液。 结果 该方法在5 min内完成数百份植物样品的DNA提取,所得DNA稀释5‒20倍即可直接用于PCR模板使用。以油菜(Brassica napus)和水稻(Oryza sativa)幼叶为材料提取DNA,成功扩增出2 000 bp以内的目的片段,扩增条带清晰、特异性良好。应用该方法进行分子标记分析,分型结果准确,重复性高,并成功用于构建油菜的局部遗传连锁图谱。 结论 建立一种“快速‒安全‒高通量”的植物DNA提取策略,简化了传统DNA提取流程,在保证扩增效果和分型准确性的基础上,大幅提升了实验效率,尤其适用于大规模植物样品的基因型检测,为分子育种和种质资源研究提供了高效、低成本的技术解决方案。
杨洋, 刘慧敏, 林俐, 王幼平, 吴健. 一种高通量快速提取植物基因组DNA的方法[J]. 生物技术通报, 2025, 41(10): 156-163.
YANG Yang, LIU Hui-min, LIN Li, WANG You-ping, WU Jian. A High-throughput and Rapid Method for Plant Genomic DNA Extraction[J]. Biotechnology Bulletin, 2025, 41(10): 156-163.
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Amplicon size (bp) |
|---|---|---|---|
| BnaC06.IDA | CAAACCAGATTCCCATTTCC | GGAATGGGAACACCTTTGGG | 514 |
| BnaA07.IDA | AAGCGAAAAGATAAGCTTGC | CGGATGATGTGTGATGCTGGA | 974 |
| ProBnaC06.IDA | GGGTGAACCCAAGAATAAC | TCGCCGCCAGAAACAGAACC | 1 215 |
| ProBnaA07.IDA | CTTGGGGCAACCCTAATCTG | CACGGAGCCATTTGGTAAT | 1 634 |
| ProBnaC04.IDA | CCACCACAAACGAGAAGACG | CACGGAGCCATTTGGTAATG | 1 803 |
| OsActin | GGTGTCATGGTCGGAATGGG | GTCCAGGGCGATGTAGGAAA | 792 |
| InDel205 | TTAAGCCCAAAACAAATTCAAATGGACTAAAA | CAAAATTTGGTTGCATAACCCTATTTCTCTTT | 127 |
| InDel348 | GAAAATAAAATTGGGAGTGGGAGGGAAA | TTTTCTCTAACGTCTCCACACGAAGATTAT | 130 |
| InDel299 | GAAACTAACTAATTTCACTTTCTCGCTCATCT | CTTTGATACTGTTGAACTAAACCGACTTTTCT | 101 |
| InDel322 | CACCCACAAACCAAAAACTAAAATTATACGAA | GCTGTAGTCTCGGTATCTCAGATTGTG | 116 |
| InDel204 | GAAGAAGCTTCGAATTTGACGGTGAC | GTCATTATTTTATGTATTTCATCCCGGGCG | 108 |
| InDel295 | AAAGCTGGCACGTCACAATATCAAT | CGAGCTATCTTTTGTTTTGGTGATATATCCTT | 130 |
| InDel319 | AGTTTTGAGTCGTATCTGAAATAAACAAAAGA | AGCACTACTTGTCGGAAAATAAAATAACATGA | 130 |
| InDel285 | TAAAAATCATTATGCAGACCGTAACTGGTAAT | ATGATGAAAATCTAAAGTGTTTTCCGTCAAAG | 106 |
| InDel620 | AGGAAAACCCCCAAAAGTAAATTTAAAAACAT | TATTGATTTTCCTCCACTGATTTGGTTTTCTA | 104 |
| InDel599 | CCATTGTAAACATCCTACGTATTGATTACTGA | CAAGCTTGTGAAAAGAAAGAAGAAGAAAATGT | 116 |
| InDel605 | GACTGATACCCACATGTATTAGAAGCAATGAC | GACTTTTACCCGGAACTGTTTACTTGAA | 119 |
表1 本研究中所用的引物序列
Table 1 Primer sequences used in this study
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Amplicon size (bp) |
|---|---|---|---|
| BnaC06.IDA | CAAACCAGATTCCCATTTCC | GGAATGGGAACACCTTTGGG | 514 |
| BnaA07.IDA | AAGCGAAAAGATAAGCTTGC | CGGATGATGTGTGATGCTGGA | 974 |
| ProBnaC06.IDA | GGGTGAACCCAAGAATAAC | TCGCCGCCAGAAACAGAACC | 1 215 |
| ProBnaA07.IDA | CTTGGGGCAACCCTAATCTG | CACGGAGCCATTTGGTAAT | 1 634 |
| ProBnaC04.IDA | CCACCACAAACGAGAAGACG | CACGGAGCCATTTGGTAATG | 1 803 |
| OsActin | GGTGTCATGGTCGGAATGGG | GTCCAGGGCGATGTAGGAAA | 792 |
| InDel205 | TTAAGCCCAAAACAAATTCAAATGGACTAAAA | CAAAATTTGGTTGCATAACCCTATTTCTCTTT | 127 |
| InDel348 | GAAAATAAAATTGGGAGTGGGAGGGAAA | TTTTCTCTAACGTCTCCACACGAAGATTAT | 130 |
| InDel299 | GAAACTAACTAATTTCACTTTCTCGCTCATCT | CTTTGATACTGTTGAACTAAACCGACTTTTCT | 101 |
| InDel322 | CACCCACAAACCAAAAACTAAAATTATACGAA | GCTGTAGTCTCGGTATCTCAGATTGTG | 116 |
| InDel204 | GAAGAAGCTTCGAATTTGACGGTGAC | GTCATTATTTTATGTATTTCATCCCGGGCG | 108 |
| InDel295 | AAAGCTGGCACGTCACAATATCAAT | CGAGCTATCTTTTGTTTTGGTGATATATCCTT | 130 |
| InDel319 | AGTTTTGAGTCGTATCTGAAATAAACAAAAGA | AGCACTACTTGTCGGAAAATAAAATAACATGA | 130 |
| InDel285 | TAAAAATCATTATGCAGACCGTAACTGGTAAT | ATGATGAAAATCTAAAGTGTTTTCCGTCAAAG | 106 |
| InDel620 | AGGAAAACCCCCAAAAGTAAATTTAAAAACAT | TATTGATTTTCCTCCACTGATTTGGTTTTCTA | 104 |
| InDel599 | CCATTGTAAACATCCTACGTATTGATTACTGA | CAAGCTTGTGAAAAGAAAGAAGAAGAAAATGT | 116 |
| InDel605 | GACTGATACCCACATGTATTAGAAGCAATGAC | GACTTTTACCCGGAACTGTTTACTTGAA | 119 |
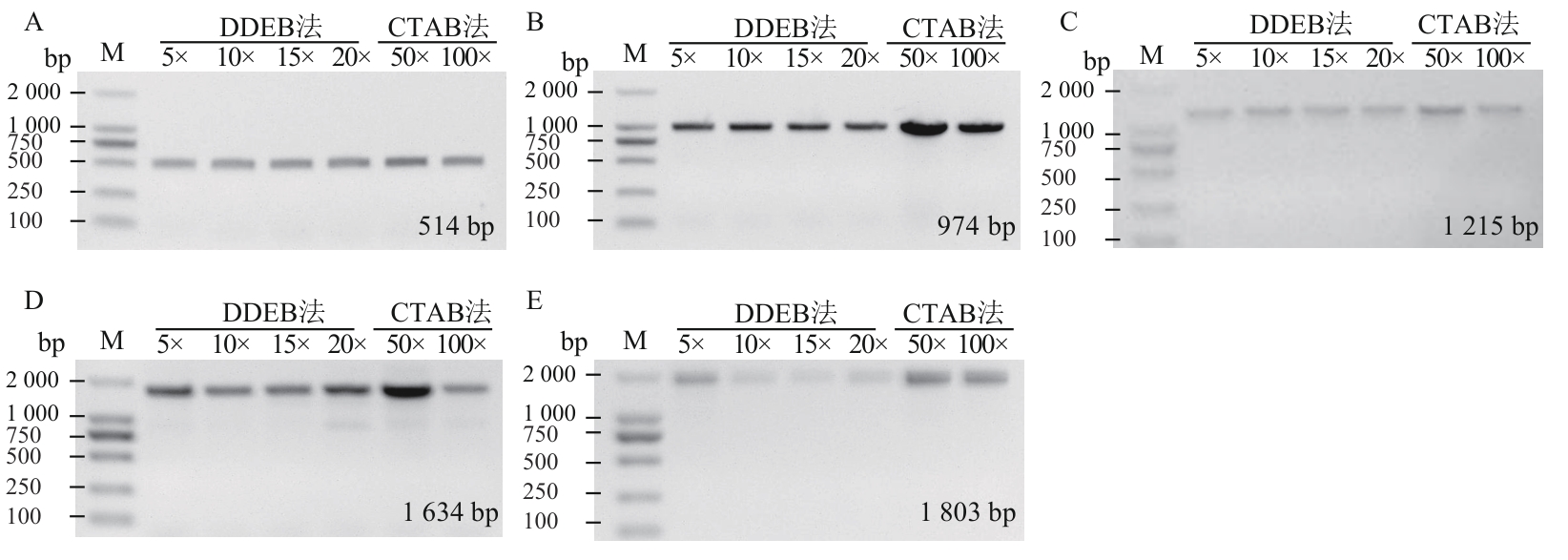
图2 DDEB法和CTAB法制备的油菜基因组DNA在不同稀释浓度下的PCR扩增效果比较A‒E扩增大小分别为514 bp(A)、974 bp(B)、1 215 bp(C)、1 634 bp(D)、1 803 bp(E);M:DNA marker
Fig. 2 Comparative PCR amplification efficiency of rapeseed genomic DNA extracted by DDEB and CTAB methods across varying dilution gradientsA‒E the amplified fragment sizes are 514 bp (A), 974 bp (B), 1 215 bp (C), 1 634 bp (D), and 1 803 bp (E), respectively; M: DNA marker
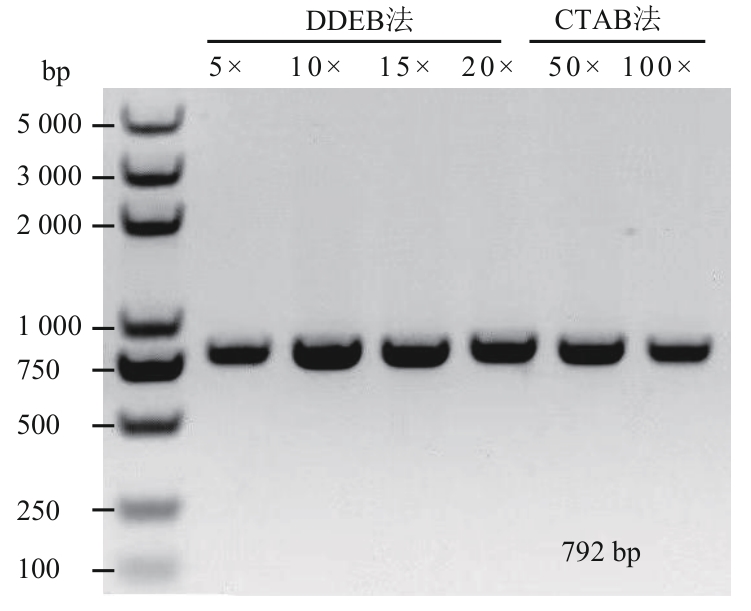
图3 DDEB法和CTAB法制备的水稻基因组DNA在不同稀释浓度下的PCR扩增效果比较
Fig. 3 Comparative PCR amplification efficiency of rice genomic DNA extracted by DDEB and CTAB methods across varying dilution gradients
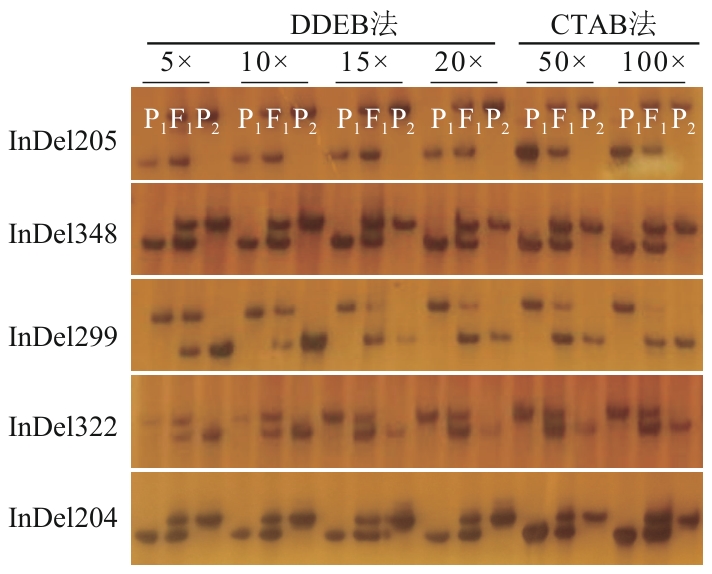
图4 DDEB法和CTAB法制备的油菜基因组DNA在InDel分子标记检测中的效果比较P1:皖油29号;P2:SWU66;F1:‘皖油29’为母本,‘SWU66’为父本的杂交一代
Fig. 4 Comparative evaluation of DDEB and CTAB methods for rapeseed genomic DNA preparation in InDel marker analysisP1: Wanyou 29; P2: SWU66; F1: the hybrid of 'Wanyou 29' (female parent)×'SWU66' (male parent)
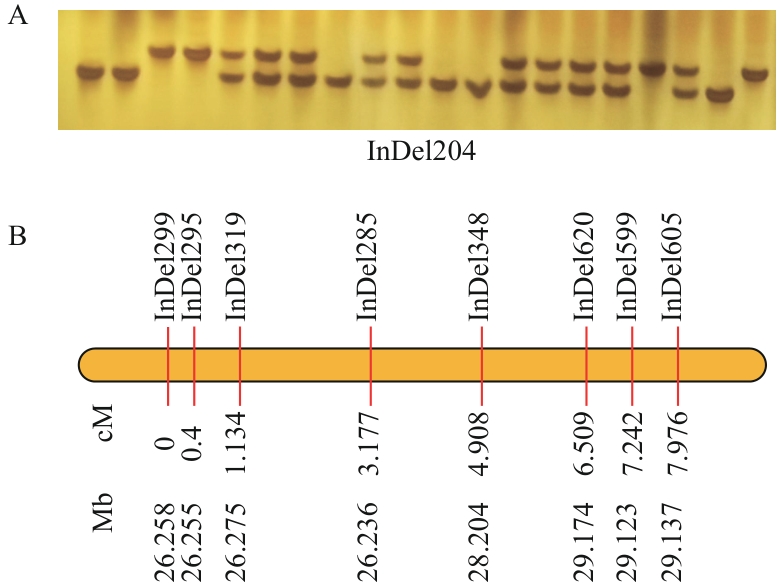
图5 基于DDEB法提取的油菜基因组DNA构建的局部遗传连锁图谱分析A:F₂群体InDel204标记基因型检测结果(聚丙烯酰胺凝胶电泳图);B:局部遗传连锁图谱,图示从上至下依次为标记名称、遗传距离(cM)及对应物理位置(Mb)
Fig. 5 Analysis of partial genetic linkage map constructed using rapeseed genomic DNA extracted by the DDEB methodA: Genotyping results of InDel204 marker in F₂ population (polyacrylamide gel electrophoresis image). B: Partial genetic linkage map showing (from top to bottom): marker names, genetic distances (cM), and corresponding physical positions (Mb)
| [1] | Amiteye S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding [J]. Heliyon, 2021, 7(10): e08093. |
| [2] | Su XM, Lyu HJ, Li J, et al. Map-based cloning and characterization of yg-2, a gene conferring yellow-green leaf in tomato (Solanum lycopersicum) [J]. Mol Breed, 2024, 44(12): 81. |
| [3] | Wang JC, Chen YJ, Zou Q. Comparative genomics and functional genomics analysis in plants [J]. Int J Mol Sci, 2023, 24(7): 6539. |
| [4] | Ben-Amar A, Mliki A. Timely gene detection assay and reliable screening of genetically engineered plants using an improved direct PCR-based technology [J]. Transgenic Res, 2021, 30(3): 263-274. |
| [5] | Holst-Jensen A, Rønning SB, Løvseth A, et al. PCR technology for screening and quantification of genetically modified organisms (GMOs) [J]. Anal Bioanal Chem, 2003, 375(8): 985-993. |
| [6] | Avenot HF, Jaime-Frias R, Travadon R, et al. Development of PCR-based assays for rapid and reliable detection and identification of canker-causing pathogens from symptomatic almond trees [J]. Phytopathology, 2022, 112(8): 1710-1722. |
| [7] | Komori T, Nitta N. A simple method to control the seed purity of Japonica hybrid rice varieties using PCR-based markers [J]. Plant Breed, 2004, 123(6): 549-553. |
| [8] | Stewart CN Jr, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications [J]. Biotechniques, 1993, 14(5): 748-750. |
| [9] | Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components[J]. Plant Mol Biol Rep, 1997, 15(1): 8-15. |
| [10] | Allen GC, Flores-Vergara MA, Krasynanski S, et al. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide [J]. Nat Protoc, 2006, 1(5): 2320-2325. |
| [11] | Feliciello I, Chinali G. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia coli [J]. Anal Biochem, 1993, 212(2): 394-401. |
| [12] | Xia YM, Chen FS, Du Y, et al. A modified SDS-based DNA extraction method from raw soybean [J]. Biosci Rep, 2019, 39(2): BSR20182271. |
| [13] | Werner O, Ros RM, Guerra J. Direct amplification and NaOH extraction: two rapid and simple methods for preparing bryophyte DNA for polymerase chain reaction (PCR) [J]. J Bryol, 2002, 24(2): 127-131. |
| [14] | Putri ND, Prayekti E. Optimalization of NaOH concentration in alkaline lysis method on quality and quantity of Candida albicans DNA [J]. Journal of Indonesian Medical Laboratory and Science, 2024, 5(2): 78-87. |
| [15] | 赵金艳, 刘榜, 王文君, 等. 一种适于PCR扩增的快速提取猪基因组DNA的碱裂解法 [J]. 华中农业大学学报, 2017, 36(4): 90-94. |
| Zhao JY, Liu B, Wang WJ, et al. A rapid alkaline lysis method of extracting pig genomic DNA for PCR amplification [J]. J Huazhong Agric Univ, 2017, 36(4): 90-94. | |
| [16] | 曹文波, 郑璐璐, 谢文海. 一种提取植物基因组DNA的方法——改良尿素法 [J]. 华中师范大学学报: 自然科学版, 2008, 42(3): 448-451. |
| Cao WB, Zheng LL, Xie WH. The modification urea method: an improved method for plant DNA isolation [J]. J Huazhong Norm Univ Nat Sci, 2008, 42(3): 448-451. | |
| [17] | 穆春华, 张发军, 李文才, 等. 玉米叶片基因组快速提取方法研究 [J]. 玉米科学, 2010, 18(3): 170-172. |
| Mu CH, Zhang FJ, Li WC, et al. A method of genomic DNA extraction of maize [J]. J Maize Sci, 2010, 18(3): 170-172. | |
| [18] | 肖帅, 李鳞霞, 李海鸥. 一种高效的植物DNA提取和PCR扩增体系建立 [J]. 亚热带植物科学, 2018, 47(3): 207-210. |
| Xiao S, Li LX, Li HO. An efficient plant DNA extraction and PCR amplification system [J]. Subtrop Plant Sci, 2018, 47(3): 207-210. | |
| [19] | 陈欲, 汪小福, 陈笑芸, 等. 基因组DNA快速提取及在转基因大豆检测中的应用 [J]. 中国油料作物学报, 2021, 43(1): 70-76. |
| Chen Y, Wang XF, Chen XY, et al. Rapid extraction of genomic DNA and its application in detection of genetically modified soybeans [J]. Chin J Oil Crop Sci, 2021, 43(1): 70-76. | |
| [20] | 金素奎, 国倩倩, 刘巧泉, 等. 一种水稻叶片基因组DNA简易提取方法 [J]. 生物技术通报, 2025, 41(1): 74-84. |
| Jin SK, Guo QQ, Liu QQ, et al. A simplified method for extracting genomic DNA from rice leaves [J]. Biotechnol Bull, 2025, 41(1): 74-84. | |
| [21] | 王慧娜, 初志战, 马兴亮, 等. 高通量PCR模板植物基因组DNA制备方法 [J]. 作物学报, 2013, 39(7): 1200-1205. |
| Wang HN, Chu ZZ, Ma XL, et al. A high through-put protocol of plant genomic DNA preparation for PCR [J]. Acta Agron Sin, 2013, 39(7): 1200-1205. | |
| [22] | Ahmed I, Islam M, Arshad W, et al. High-quality plant DNA extraction for PCR: an easy approach [J]. J Appl Genet, 2009, 50(2): 105-107. |
| [23] | Bellstedt DU, Pirie MD, Visser JC, et al. A rapid and inexpensive method for the direct PCR amplification of DNA from plants [J]. Am J Bot, 2010, 97(7): e65-e68. |
| [24] | Hwang H, Bae SC, Lee S, et al. A rapid and simple genotyping method for various plants by direct-PCR [J]. Plant Breed Biotech, 2013, 1(3): 290-297. |
| [25] | Sajib AA, Bhuiya MAI, Huque R. A simple, efficient and rapid method for good quality DNA extraction from rice grains [J]. Rice Sci, 2017, 24(2): 119-122. |
| [26] | Wang TY, Wang L, Zhang JH, et al. A simplified universal genomic DNA extraction protocol suitable for PCR [J]. Genet Mol Res, 2011, 10(1): 519-525. |
| [27] | Li X, Liu C, Wang DB, et al. Persistent pollution of genetic materials in a typical laboratory environment [J]. J Hazard Mater, 2024, 470: 134201. |
| [28] | Wu YH, Bhat PR, Close TJ, et al. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph [J]. PLoS Genet, 2008, 4(10): e1000212. |
| [29] | Wang BW, Liu XL, Li ZX, et al. A nuclease-dead Cas9-derived tool represses target gene expression [J]. Plant Physiol, 2024, 195(3): 1880-1892. |
| [30] | Abd Razak WNR, Bahrain KBS, Bostanudin MF, et al. Evaluation of the inhibitory effects of pharmaceutical excipients toward the efficiency of polymerase chain reaction (PCR) [J]. Int J Pharm Res, 2020, 12(3): 1968-1976. |
| [1] | 刘佳, 任义尚, 荆晓艳, 许拉. 基于拉曼光谱“先筛后养”策略在功能微生物资源挖掘中的应用与展望[J]. 生物技术通报, 2026, 42(5): 1-13. |
| [2] | 王芳, 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍, 姜亮. 植物和细菌TurboID邻近蛋白标记方法的建立[J]. 生物技术通报, 2025, 41(9): 44-53. |
| [3] | 邓美壁, 严浪, 詹志田, 朱敏, 和玉兵. RUBY辅助的水稻高效CRISPR基因编辑[J]. 生物技术通报, 2025, 41(8): 65-73. |
| [4] | 侯鹰翔, 费思恬, 黎妮, 李兰, 宋松泉, 王伟平, 张超. 水稻miRNAs响应生物胁迫研究进展[J]. 生物技术通报, 2025, 41(7): 69-80. |
| [5] | 吴浩, 董伟峰, 贺子天, 李艳肖, 谢辉, 孙明哲, 沈阳, 孙晓丽. 水稻BXL基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(6): 87-98. |
| [6] | 周志国, 樊双虎, 邓晨, 冯雪. 2,4-表油菜素内酯对镉胁迫下胡萝卜幼苗生理特性的影响[J]. 生物技术通报, 2025, 41(5): 165-174. |
| [7] | 杜量衡, 唐黄磊, 张治国. 控制水稻光响应基因ELM1的图位克隆[J]. 生物技术通报, 2025, 41(5): 82-89. |
| [8] | 刘园园, 陈析丰, 钱前, 高振宇. 水稻穗发育调控的分子机制研究进展[J]. 生物技术通报, 2025, 41(5): 1-13. |
| [9] | 侯亚涛, 李迎辉, 邓磊, 李常保, 李传友, 孙传龙. 番茄果重基因功能型分子标记的开发及群体基因型分析[J]. 生物技术通报, 2025, 41(4): 98-105. |
| [10] | 陈晓军, 惠建, 马洪文, 白海波, 钟楠, 李稼润, 樊云芳. 利用单碱基基因编辑技术创制OsALS抗除草剂水稻种质资源[J]. 生物技术通报, 2025, 41(4): 106-114. |
| [11] | 夏馨媛, 薛道晟, 李鑫静, 龙俊杰, 陆开形, 丁沃娜, 李梦莎. 稻油轮作土壤多功能促生菌的鉴定及其对油菜生长和根际细菌群落的影响[J]. 生物技术通报, 2025, 41(4): 289-301. |
| [12] | 王琛, 刘国梅, 陈畅, 张晋龙, 姚琳, 孙璇, 杜春芳. 白菜型油菜CCDs家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(3): 161-170. |
| [13] | 李欣芃, 张武汉, 张莉, 舒服, 何强, 郭杨, 邓华凤, 王悦, 孙平勇. γ射线诱变创制水稻突变体及其分子鉴定[J]. 生物技术通报, 2025, 41(3): 35-43. |
| [14] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| [15] | 范艳飞, 叶露幻, 李雨桐, 王钏跞, 张瑞, 罗建华, 王鹏. 第二十七届中国科协年会学术论文运用小麦杂交株系挖掘环式光合电子传递调控基因并应用于作物高光效改造[J]. 生物技术通报, 2025, 41(10): 72-86. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||