








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (5): 175-182.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1039
Previous Articles Next Articles
LI Yu1( ), CHEN Gang1(
), CHEN Gang1( ), MA Qian1(
), MA Qian1( ), KUANG Jie-hua1, CHEN You-ming2
), KUANG Jie-hua1, CHEN You-ming2
Received:2021-08-16
Online:2022-05-26
Published:2022-06-10
Contact:
CHEN Gang,MA Qian
E-mail:1948463705@qq.com;cheng@gdou.edu.cn;maq@gdou.edu.cn
LI Yu, CHEN Gang, MA Qian, KUANG Jie-hua, CHEN You-ming. Cloning of nanos1 Gene of Rachycentron canadum and Its Expression Analysis in the Embryonic and Gonadal Development[J]. Biotechnology Bulletin, 2022, 38(5): 175-182.
| 引物名称 Primer | 序列Sequence(5'-3') | 用途 Usage |
|---|---|---|
| Rcnanos1-F | ATGGATTTTCTCAACCACAACTATT | CDS序列克隆 CDS sequence cloning |
| Rcnanos1-R | TTAGAATATTTTCATCCTCTTACCACC | |
| Rcnanos1-3' F1 | GCCCAAAATCTGCGTCTTCTGC | 3' 末端序列克隆 3' RACE |
| Rcnanos1-3' F2 | TCAAAAGACCAGCCATCCCAGC | |
| Rcnanos1-5' R1 | GCTCCTGGAAGCGGGTTTGC | 5' 末端序列克隆 5' RACE |
| Rcnanos1-5' R2 | CGATTACGCACGGACACGCC | |
| Rcnanos1-F1 | AGAACCCCAACTCCATCACC | Rcnanos1的qRT-PCR检测 Expression ofRcnanos1 |
| Rcnanos1-R1 | TCTGGAAGGGGCTCAGTATC | |
| β-actin-F | AGGGAAATTGTGCGTGAC | 内参基因 Internal control gene |
| β-actin-R | AGGCAGCTCGTAGCTCTT |
Table 1 Primer sequences used in this study
| 引物名称 Primer | 序列Sequence(5'-3') | 用途 Usage |
|---|---|---|
| Rcnanos1-F | ATGGATTTTCTCAACCACAACTATT | CDS序列克隆 CDS sequence cloning |
| Rcnanos1-R | TTAGAATATTTTCATCCTCTTACCACC | |
| Rcnanos1-3' F1 | GCCCAAAATCTGCGTCTTCTGC | 3' 末端序列克隆 3' RACE |
| Rcnanos1-3' F2 | TCAAAAGACCAGCCATCCCAGC | |
| Rcnanos1-5' R1 | GCTCCTGGAAGCGGGTTTGC | 5' 末端序列克隆 5' RACE |
| Rcnanos1-5' R2 | CGATTACGCACGGACACGCC | |
| Rcnanos1-F1 | AGAACCCCAACTCCATCACC | Rcnanos1的qRT-PCR检测 Expression ofRcnanos1 |
| Rcnanos1-R1 | TCTGGAAGGGGCTCAGTATC | |
| β-actin-F | AGGGAAATTGTGCGTGAC | 内参基因 Internal control gene |
| β-actin-R | AGGCAGCTCGTAGCTCTT |

Fig.1 Complete sequence of Rcnanos1 cDNA and analysis of deduced amino acid sequence The initiation codon ATG and stop codon TAA are in bold. The polyadenylation signal ATTAAA are in bold and italic. The zinc finger motif are highlighted in grey shadow
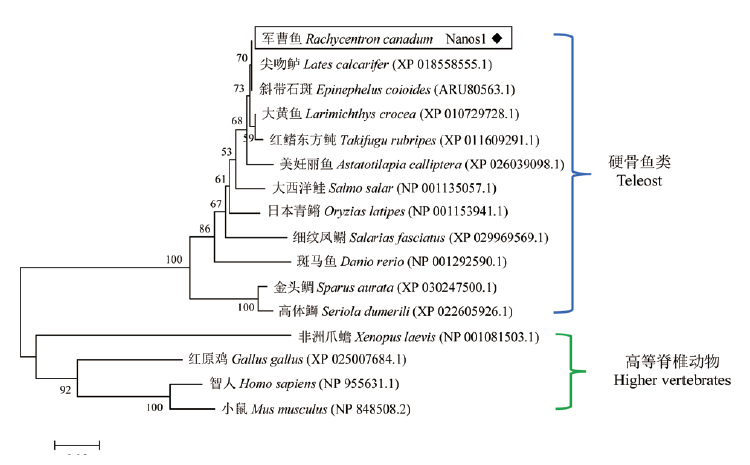
Fig.3 Phylogenetic tree of nanos1 amino acid sequences based on Neighbor-Joining(NJ)method Numbers indicates the confidence level of each branch,the scale bar 0.05 in terms of genetic distance is indicated below the tree

Fig.4 Expressions of Rcnanos1 in the tissues of R. canadum by qRT-PCR LI:Liver. SP:Spleen. KI:Kidney. BR:Brain. HE:Heart. GI:Gill. TE:Testis. OV:Ovary. ST:Stomach. IN:Intestines. MU:Muscle. SK:Skin. EY:Eye. Different superscripts indicate significant difference(P<0.05)
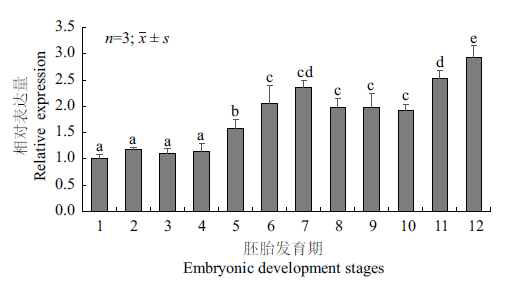
Fig.5 Expressions of Rcnanos1 during the embryonic deve-lopment stages 1:1-cell stage;2:2-cell stage;3:4-cell stage;4:8-cell stage;5:16-cell stage;6:poly-cell stage;7:blastocyst stage;8:gastrulation stage;9:neuroblast stage;10:organ formation stage;11:hatching stage;12:1 day after hatching. Different superscripts indicate significant difference(P<0.05)
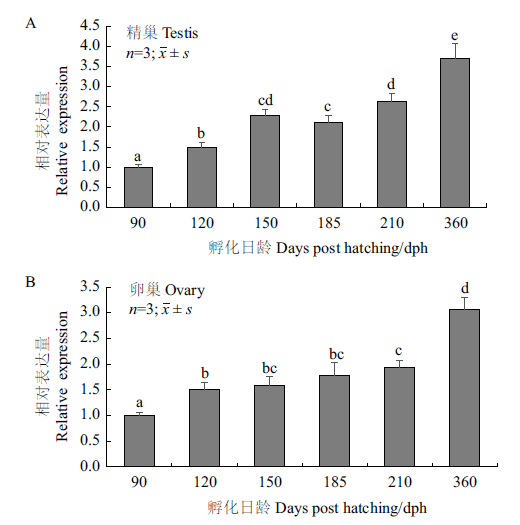
Fig.6 Rcnanos1 mRNA expression in annual gonadal deve-lopment of R. Canadum A:Rcnanos1 mRNA expression in testis;B:Rcnanos1 mRNA expression in ovary. Different superscripts indicate significant difference(P<0.05)
| [1] | 程琳, 黄天晴, 刘晨斌, 等. 鱼类原始生殖细胞标记基因研究进展[J]. 水产学杂志, 2020, 33(6):80-88. |
| Cheng L, Huang TQ, Liu CB, et al. Research perspectives:marker genes of primordial germ cells in fishes[J]. Chin J Fish, 2020, 33(6):80-88. | |
| [2] |
Xu H, Gui J, Hong Y. Differential expression of Vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp(Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate[J]. Dev Dyn, 2005, 233(3):872-882.
doi: 10.1002/dvdy.20410 URL |
| [3] |
Liu L, Hong N, Xu H, et al. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells[J]. Gene Expr Patterns, 2009, 9(7):541-548.
doi: 10.1016/j.gep.2009.06.008 URL |
| [4] |
Sun ZH, Wang Y, Lu WJ, et al. Divergent expression patterns and function implications of four Nanos genes in a hermaphroditic fish, Epinephelus coioides[J]. Int J Mol Sci, 2017, 18(4):685.
doi: 10.3390/ijms18040685 URL |
| [5] |
Bhat N, Hong Y. Cloning and expression of boule and dazl in the Nile tilapia(Oreochromis niloticus)[J]. Gene, 2014, 540(2):140-145.
doi: 10.1016/j.gene.2014.02.057 URL |
| [6] |
Shen R, Xie T. NANOS:a germline stem cell’s Guardian Angel[J]. J Mol Cell Biol, 2010, 2(2):76-77.
doi: 10.1093/jmcb/mjp043 pmid: 20008335 |
| [7] |
Ye H, Chen X, Wei Q, et al. Molecular and expression characterization of a nanos1 homologue in Chinese sturgeon, Acipenser sinensis[J]. Gene, 2012, 511(2):285-292.
doi: 10.1016/j.gene.2012.09.005 URL |
| [8] |
Kadyrova LY, Habara Y, Lee TH, et al. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline[J]. Development, 2007, 134(8):1519-1527.
doi: 10.1242/dev.002212 URL |
| [9] |
Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation[J]. Science, 2004, 303(5666):2016-2019.
doi: 10.1126/science.1093983 URL |
| [10] | Tan XG, Sui YL, Li MJ, et al. Characterization of nanos1 homolog in the olive flounder, Paralichthys olivaceus(temminck and Schlegel, 1846)[J]. Turk J Fish Aquat Sci, 2020, 20(6):421-429. |
| [11] |
Haraguchi S, Tsuda M, Kitajima S, et al. nanos1:a mouse Nanos gene expressed in the central nervous system is dispensable for normal development[J]. Mech Dev, 2003, 120(6):721-731.
doi: 10.1016/S0925-4773(03)00043-1 URL |
| [12] | 肖懿哲, 朱友芳, 等. 施氏鲟asnanos1基因序列分析及其在雌雄不同组织中表达[J]. 海洋科学, 2017, 41(4):17-23. |
| Xiao YZ, Zhu YF, et al. Sequence and differential expression analyses of the Asnanos1 gene in different tissues of male and female Acipenser schrenckii[J]. Mar Sci, 2017, 41(4):17-23. | |
| [13] |
Köprunner M, Thisse C, Thisse B, et al. A zebrafish Nanos-related gene is essential for the development of primordial germ cells[J]. Genes Dev, 2001, 15(21):2877-2885.
doi: 10.1101/gad.212401 URL |
| [14] |
Zhu W, Wang T, Zhao C, et al. Evolutionary conservation and divergence of Vasa, Dazl and Nanos1 during embryogenesis and gametogenesis in dark sleeper(Odontobutis potamophila)[J]. Gene, 2018, 672:21-33.
doi: 10.1016/j.gene.2018.06.016 URL |
| [15] |
Han K, Chen S, Cai M, et al. Nanos3 not nanos1 and nanos2 is a germ cell marker gene in large yellow croaker during embryogenesis[J]. Comp Biochem Physiol B Biochem Mol Biol, 2018, 218:13-22.
doi: 10.1016/j.cbpb.2018.01.002 URL |
| [16] | 苏敏, 陈元仲, 吕博彦, 等. nanos同源基因在黑脊倒刺鲃生殖细胞中的表达研究[J]. 生物技术, 2012, 22(4):28-32. |
| Su M, et al. Expression of Nanos homolog gene in germ cells of Spinibarbus caldwelli[J]. Biotechnology, 2012, 22(4):28-32. | |
| [17] | Sajeevan MK, Kurup BM. Fecundity and spawning frequency of Cobia, Rachycentron canadum(Linnaeus, 1766)from the North West coast of India[J]. Indian journal of marine sciences, 2016, 45(8):933-936. |
| [18] |
Darden TL, Robinson JD, Strand AE, et al. Forecasting the genetic impacts of net pen failures on gulf of Mexico cobia populations using individual-based model simulations[J]. J World Aquacult Soc, 2017, 48(1):20-34.
doi: 10.1111/jwas.12333 URL |
| [19] | Nguyen MV, Thi Phan LM. Influences of bleeding conditions on the quality and lipid degradation of cobia(Rachycentron canadum)fillets during frozen storage[J]. Turk J Fish and Aquat Sci, 2018, 18(2):289-300. |
| [20] |
Kuo CH, Liao HZ, et al. Highly efficient extraction of EPA/DHA-enriched oil from cobia liver using homogenization plus sonication[J]. Eur J Lipid Sci Technol, 2017, 119(10):1600466.
doi: 10.1002/ejlt.201600466 URL |
| [21] | 刘筠. 中国养殖鱼类繁殖生理学[M]. 北京: 农业出版社, 1993:20-42. |
| Liu Y. Propagation physiology of main cultivated fish in China[M]. Beijing: Agriculture Press, 1993:20-42. | |
| [22] |
Gribouval L, Sourdaine P, et al. The nanos1 gene was duplicated in early Vertebrates and the two paralogs show different gonadal expression profiles in a shark[J]. Sci Rep, 2018, 8(1):6942.
doi: 10.1038/s41598-018-24643-1 pmid: 29720681 |
| [23] |
De Keuckelaere E, et al. Nanos genes and their role in development and beyond[J]. Cell Mol Life Sci, 2018, 75(11):1929-1946.
doi: 10.1007/s00018-018-2766-3 pmid: 29397397 |
| [24] | Lai F. An investigation of Nanos1 function during primordial germ cell development in Xenopus laevis[D]. University of Miami, 2012. |
| [25] |
Hashimoto H, Hara K, Hishiki A, et al. Crystal structure of zinc-finger domain of Nanos and its functional implications[J]. EMBO Rep, 2010, 11(11):848-853.
doi: 10.1038/embor.2010.155 pmid: 20948543 |
| [26] |
Ye B, Petritsch C, Clark IE, et al. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons[J]. Curr Biol, 2004, 14(4):314-321.
doi: 10.1016/j.cub.2004.01.052 URL |
| [27] | 徐钢春, 鲍明明, 等. 鱼类性腺发育及产卵类型研究进展[J]. 长江大学学报:自然科学版, 2017, 14(6):43-48. |
| Xu GC, Bao MM, et al. Research progress on gonadal development and spawning types in fish[J]. J Yangtze Univ:Nat Sci Ed, 2017, 14(6):43-48. | |
| [28] |
Viotti M, Nowotschin S, Hadjantonakis AK. SOX 17 links gut endoderm morphogenesis and germ layer segregation[J]. Nat Cell Biol, 2014, 16(12):1146-1156.
doi: 10.1038/ncb3070 URL |
| [29] | 宋卉, 王树迎. 鱼类原始生殖细胞的研究进展[J]. 动物医学进展, 2004, 25(5):22-23. |
| Song H, Wang SY. Progress in fish primordial germ cells[J]. Prog Vet Med, 2004, 25(5):22-23. |
| [1] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [2] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [3] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [4] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [5] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [6] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [7] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [8] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [9] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [10] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [11] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [12] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [13] | YOU Chui-huai, XIE Jin-jin, ZHANG Ting, CUI Tian-zhen, SUN Xin-lu, ZANG Shou-jian, WU Yi-ning, SUN Meng-yao, QUE You-xiong, SU Ya-chun. Identification of the Lipoxygenase Gene GeLOX1 and Expression Analysis Under Low Temperature Stress in Gelsmium elegans [J]. Biotechnology Bulletin, 2023, 39(11): 318-327. |
| [14] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| [15] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||