








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 313-324.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0748
ZHANG Dan-dan1( ), ZHAO Rui-xue1,2(
), ZHAO Rui-xue1,2( ), XIAN Guo-jian1,3, XIONG He1
), XIAN Guo-jian1,3, XIONG He1
Received:2023-08-05
Online:2024-02-26
Published:2024-03-13
Contact:
ZHAO Rui-xue
E-mail:zhangdandan01@caas.cn;zhaoruixue@caas.cn
ZHANG Dan-dan, ZHAO Rui-xue, XIAN Guo-jian, XIONG He. Trait-regulated-genes Ontology Model Construction and Application by Integrating Cross-species Scientific Data[J]. Biotechnology Bulletin, 2024, 40(2): 313-324.
| 数据类型维度 Data type dimension | 实体名称 Entity name | 数据属性 Data attribute |
|---|---|---|
| 基因水平 Gene level | 基因 Gene | 基因标识符;物种;物理位置;转录本名称;PANTHER注释编号 Gene identity; species; location; transcript name; PANTHER identity |
| 基因符号 Gene symbol | 功能描述 Function description | |
| 蛋白水平 Protein level | 蛋白 Protein | 蛋白标识符;物种;首次被发现时间;功能描述;影响表型描述;文献编号 Protein identity; species; date of creation; function description; phenotype disruption; PubMed identity |
| 蛋白家族 Protein family | 蛋白家族数据库编号;名称 Pfam identity; name | |
| 酶 Enzyme | EC编号;名称 EC number; name | |
| 结构域 Domain | 名称;类型 Name; type | |
| 亚细胞定位 Subcellular localization | 名称;类型 Name; type | |
| 富集通路水平 Enrichment pathways level | 生物学过程 Biological process | GO编号;名称 GO identity; name |
| 细胞组分 Cellular component | GO编号;名称 GO identity; name | |
| 分子功能 Molecular function | GO编号;名称 GO identity; name | |
| 代谢通路 Metabolic pathway | KO编号;名称 KO identity; name | |
| 信号通路 Signal pathway | 名称;类型 Name; type | |
| 性状水平 Trait level | 性状 Trait | 名称;类型 Name; type |
Table 1 Description of entity classes in trait-regulated-genes ontology model
| 数据类型维度 Data type dimension | 实体名称 Entity name | 数据属性 Data attribute |
|---|---|---|
| 基因水平 Gene level | 基因 Gene | 基因标识符;物种;物理位置;转录本名称;PANTHER注释编号 Gene identity; species; location; transcript name; PANTHER identity |
| 基因符号 Gene symbol | 功能描述 Function description | |
| 蛋白水平 Protein level | 蛋白 Protein | 蛋白标识符;物种;首次被发现时间;功能描述;影响表型描述;文献编号 Protein identity; species; date of creation; function description; phenotype disruption; PubMed identity |
| 蛋白家族 Protein family | 蛋白家族数据库编号;名称 Pfam identity; name | |
| 酶 Enzyme | EC编号;名称 EC number; name | |
| 结构域 Domain | 名称;类型 Name; type | |
| 亚细胞定位 Subcellular localization | 名称;类型 Name; type | |
| 富集通路水平 Enrichment pathways level | 生物学过程 Biological process | GO编号;名称 GO identity; name |
| 细胞组分 Cellular component | GO编号;名称 GO identity; name | |
| 分子功能 Molecular function | GO编号;名称 GO identity; name | |
| 代谢通路 Metabolic pathway | KO编号;名称 KO identity; name | |
| 信号通路 Signal pathway | 名称;类型 Name; type | |
| 性状水平 Trait level | 性状 Trait | 名称;类型 Name; type |
| 数据属性名称 Data attributes | 释义 Description |
|---|---|
| 基因标识符 Gene identity | 基因的唯一标识符用于区分基因类实例 |
| 转录本名称 Transcript name | 描述基因在转录后所对应的名称 |
| 物理位置 Location | 描述基因在染色体中的具体物理位点 |
| PANTHER数据库编号 PANTHER identity | 描述基因在PANTHER数据库中的具体注释信息编号 |
| 蛋白标识符 Protein identity | 蛋白的唯一标识符用于区分蛋白类实例 |
| 首次被发现时间 Date of creation | 描述蛋白初次被发现的时间 |
| 影响表型描述 Phenotype disruption | 描述调控表型的主要分子机制 |
| 物种 Species | 描述基因或蛋白所属于的物种 |
| 文献编号 PubMed identity | 描述报道该蛋白的文献在PubMed数据库中的编号 |
| 功能描述 Function description | 描述蛋白类实例主要的功能 |
| 蛋白家族数据库编号 Pfam identity | 描述蛋白家族在Pfam数据库中的具体编号信息 |
| EC编号 EC number | 描述基因所编码酶的类型信息所属的EC编号 |
| KO数据库编号 KO identity | 描述基因在KO数据库中的具体注释信息编号 |
| GO数据库编号 GO identity | 描述基因的分子调控、细胞组分、生物学过程信息在GO数据库中所对应的编号 |
| 名称 Name | 描述性状、蛋白家族、代谢途径等核心类的具体实例名称 |
| 类型 Type | 描述性状、结构域、通路等核心类所属的类别名称 |
Table 2 Description of data attributes in trait-regulated-genes ontology model
| 数据属性名称 Data attributes | 释义 Description |
|---|---|
| 基因标识符 Gene identity | 基因的唯一标识符用于区分基因类实例 |
| 转录本名称 Transcript name | 描述基因在转录后所对应的名称 |
| 物理位置 Location | 描述基因在染色体中的具体物理位点 |
| PANTHER数据库编号 PANTHER identity | 描述基因在PANTHER数据库中的具体注释信息编号 |
| 蛋白标识符 Protein identity | 蛋白的唯一标识符用于区分蛋白类实例 |
| 首次被发现时间 Date of creation | 描述蛋白初次被发现的时间 |
| 影响表型描述 Phenotype disruption | 描述调控表型的主要分子机制 |
| 物种 Species | 描述基因或蛋白所属于的物种 |
| 文献编号 PubMed identity | 描述报道该蛋白的文献在PubMed数据库中的编号 |
| 功能描述 Function description | 描述蛋白类实例主要的功能 |
| 蛋白家族数据库编号 Pfam identity | 描述蛋白家族在Pfam数据库中的具体编号信息 |
| EC编号 EC number | 描述基因所编码酶的类型信息所属的EC编号 |
| KO数据库编号 KO identity | 描述基因在KO数据库中的具体注释信息编号 |
| GO数据库编号 GO identity | 描述基因的分子调控、细胞组分、生物学过程信息在GO数据库中所对应的编号 |
| 名称 Name | 描述性状、蛋白家族、代谢途径等核心类的具体实例名称 |
| 类型 Type | 描述性状、结构域、通路等核心类所属的类别名称 |
| 对象属性名称Object attributes | 描述对象 Object | 数据来源 Data source | 释义 Description |
|---|---|---|---|
| 与……有关 Associates with | 蛋白;性状Protein; trait | UniProt, PubMed | 描述蛋白和性状之间的关联关系 |
| 与……同源 Homologous to | 蛋白;蛋白 Protein ; protein | UniProt | 描述蛋白与蛋白之间的同源关系 |
| 与......互作 Interacts with | 蛋白;蛋白 Protein ; protein | UniProt, STRING | 描述蛋白与蛋白间相互作用的关系 |
| 与……相对应 Corresponding to | 蛋白;基因Protein; gene | UniProt, Phytozome, Ensembl plants, RGAP | 描述基因和蛋白间的对应关系 |
| 与……一致 Identify with | 蛋白;基因符号Protein; gene symbol | UniProt | 描述蛋白和基因符号的关联关系 |
| 参与...... Involves in | 蛋白;信号通路 Protein; signal pathway | UniProt | 描述蛋白和信号通路的关联关系 |
| 表达于 Located in | 蛋白;亚细胞定位位置 Protein; subcellular localization | UniProt | 描述蛋白亚细胞定位的位置 |
| 具有......蛋白结构域 Has protein domain | 蛋白;结构域Protein; domain | UniProt | 描述蛋白所具有的蛋白结构域 |
| 属于...... Belongs to | 蛋白;蛋白家族Protein; protein family | UniProt, Pfam | 描述蛋白所属的蛋白家族 |
| 表达于 Located in | 基因;细胞组分 Gene; cellular component | Phytozome, Ensembl plants, RGAP, KEGG | 描述基因的亚细胞定位 |
| 行使功能 Performs | 基因;分子功能 Gene; molecular function | Phytozome, Ensembl plants, RGAP, GO | 描述基因在分子水平上所行使的功能 |
| 参与...... Involves in | 基因;生物学过程 Gene; biological process | Phytozome, Ensembl plants, RGAP, GO | 描述基因功能所直接参与的生物过程 |
| 参与..... Involves in | 基因;代谢通路 Gene; metabolic pathway | Phytozome, Ensembl plants, RGAP, KEGG | 描述基因在代谢通路中的功能 |
| 编码......类型的酶 Encodes the enzyme type | 基因;酶Gene; enzyme | Phytozome, Ensembl plants, RGAP, KEGG | 描述基因所编码蛋白所属的酶的类型 |
Table 3 Description of object attributes in trait-regulated-genes ontology model and data sources
| 对象属性名称Object attributes | 描述对象 Object | 数据来源 Data source | 释义 Description |
|---|---|---|---|
| 与……有关 Associates with | 蛋白;性状Protein; trait | UniProt, PubMed | 描述蛋白和性状之间的关联关系 |
| 与……同源 Homologous to | 蛋白;蛋白 Protein ; protein | UniProt | 描述蛋白与蛋白之间的同源关系 |
| 与......互作 Interacts with | 蛋白;蛋白 Protein ; protein | UniProt, STRING | 描述蛋白与蛋白间相互作用的关系 |
| 与……相对应 Corresponding to | 蛋白;基因Protein; gene | UniProt, Phytozome, Ensembl plants, RGAP | 描述基因和蛋白间的对应关系 |
| 与……一致 Identify with | 蛋白;基因符号Protein; gene symbol | UniProt | 描述蛋白和基因符号的关联关系 |
| 参与...... Involves in | 蛋白;信号通路 Protein; signal pathway | UniProt | 描述蛋白和信号通路的关联关系 |
| 表达于 Located in | 蛋白;亚细胞定位位置 Protein; subcellular localization | UniProt | 描述蛋白亚细胞定位的位置 |
| 具有......蛋白结构域 Has protein domain | 蛋白;结构域Protein; domain | UniProt | 描述蛋白所具有的蛋白结构域 |
| 属于...... Belongs to | 蛋白;蛋白家族Protein; protein family | UniProt, Pfam | 描述蛋白所属的蛋白家族 |
| 表达于 Located in | 基因;细胞组分 Gene; cellular component | Phytozome, Ensembl plants, RGAP, KEGG | 描述基因的亚细胞定位 |
| 行使功能 Performs | 基因;分子功能 Gene; molecular function | Phytozome, Ensembl plants, RGAP, GO | 描述基因在分子水平上所行使的功能 |
| 参与...... Involves in | 基因;生物学过程 Gene; biological process | Phytozome, Ensembl plants, RGAP, GO | 描述基因功能所直接参与的生物过程 |
| 参与..... Involves in | 基因;代谢通路 Gene; metabolic pathway | Phytozome, Ensembl plants, RGAP, KEGG | 描述基因在代谢通路中的功能 |
| 编码......类型的酶 Encodes the enzyme type | 基因;酶Gene; enzyme | Phytozome, Ensembl plants, RGAP, KEGG | 描述基因所编码蛋白所属的酶的类型 |
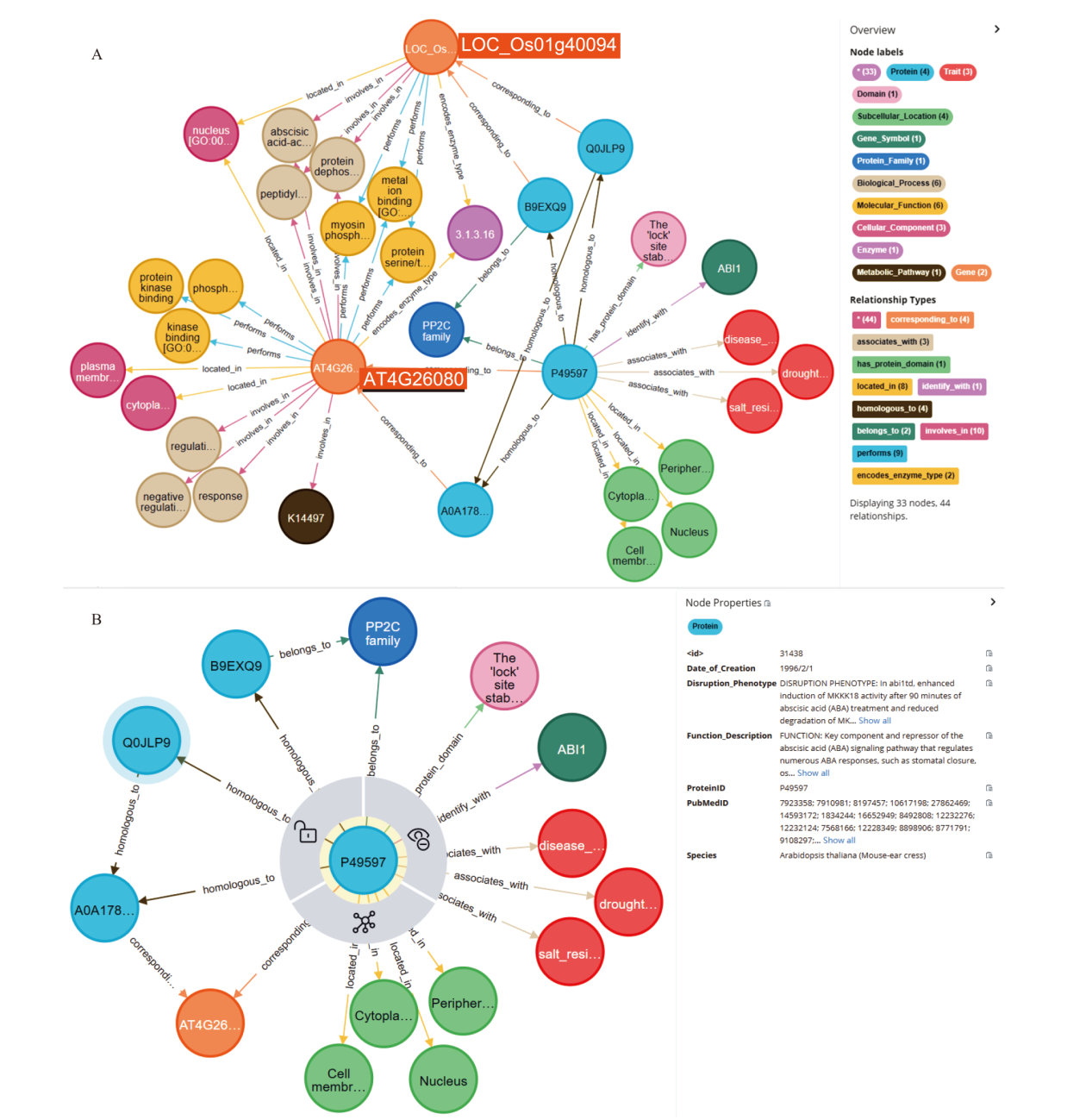
Fig. 2 Hierarchical knowledge structure of trait-regulated-genes knowledge graph A: Multi-dimensional scientific data association retrieval of gene LOC_Os01g40094. B: Data attributes of protein P49597. Different colored circles and arrow lines represent different entity types and relationship types in figure A, respectively. The numbers in parentheses indicate the number of nodes and relationships of different types. Taking node types as an example, red circles represent “trait” nodes, and the number is 3. The light blue circle represents the “protein” node, the number is 4. The orange circle represents the “gene” node, and the number is 2. For example, taking relationship types as an example, the light green arrow line represents the “has protein domain” relation type, and the number is 1. The dark green arrow line represents the “ belongs to “ relation type with a number of 2

Fig. 3 Prediction of gene-regulated- trait The circles and arrow lines in different colors in the figure represent different entity types and relationship types respectively, and the numbers in brackets represent the number of nodes and relationships of different types. Taking node types as an example, the red circle represents a “trait” node and the number is 1. The light blue circle represents the “protein” node, the number of which is 7. The orange circle represents the “gene” node, with a number of 7. Take relationship types as an example, the light green arrow line represents the “has protein domain” relationship type, the number is 6. The dark green arrow line represents the “ belongs to” relation type with a number of 7. This figure shows that there are 8 association paths between the gene “TraesCS4B02G060000” node and the trait “plant height” node

Fig. 4 Mining of the elite pleiotropy gene TraesCS3D02G078500 The circles and arrow lines in different colors in the figure represent different entity types and relationship types respectively, and the numbers in brackets represent the number of nodes and relationships of different types. Taking node types as an example, the red circle represents the trait node and the number is 5. The light blue circle represents the “protein” node, the number of which is 5. The orange circle represents the “gene” node, and the number is 5. Take relationship types as an example, the orange arrow line represents the “located in” relationship type, and the number is 5. The yellow red arrow line represents the “ corresponding to “ relationship type, and the number is 5. This figure predicts that the gene “TraesCS3D02G078500” may regulate the traits of drought resistance, salt resistance, grain weight and plant height
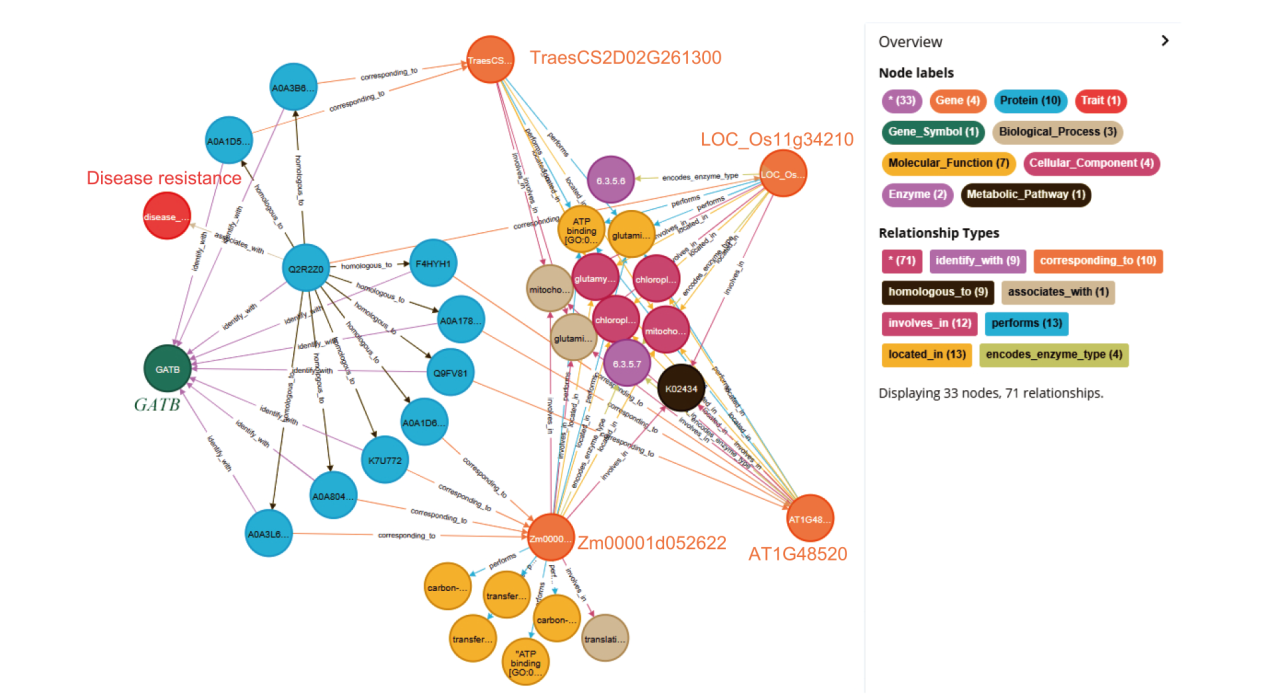
Fig. 5 Prediction of gene function across species The circles and arrow lines in different colors in the figure represent different entity types and relationship types respectively, and the numbers in brackets represent the number of nodes and relationships of different types. Taking node types as an example, the red circle represents a “trait” node and the number is 1. The light blue circle represents the “protein” node, with a number of 10. The orange circle represents the “gene” node, with a number of 4. Take relationship types as an example, the orange arrow line represents the “located in” relationship type, and the number is 13. The yellow red arrow line represents the “ corresponding to “ relationship type, and the number is 10. This figure establishes the association path between wheat gene “TraesCS2D02G261300” and Arabidopsis gene “AT1G48520”, rice gene “LOC_Os11g34210” and maize gene “Zm00001d052622”
| [1] |
Wallace JG, Rodgers-Melnick E, Buckler ES. On the road to breeding 4.0: unraveling the good, the bad, and the boring of crop quantitative genomics[J]. Annu Rev Genet, 2018, 52: 421-444.
doi: 10.1146/annurev-genet-120116-024846 pmid: 30285496 |
| [2] |
Goble C, Stevens R. State of the nation in data integration for bioinformatics[J]. J Biomed Inform, 2008, 41(5): 687-693.
doi: 10.1016/j.jbi.2008.01.008 pmid: 18358788 |
| [3] |
Hassani-Pak K, Castellote M, Esch M, et al. Developing integrated crop knowledge networks to advance candidate gene discovery[J]. Appl Transl Genom, 2016, 11: 18-26.
doi: 10.1016/j.atg.2016.10.003 pmid: 28018846 |
| [4] |
Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology[J]. Nat Genet, 2000, 25(1): 25-29.
doi: 10.1038/75556 pmid: 10802651 |
| [5] |
Zeeberg BR, Feng WM, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data[J]. Genome Biol, 2003, 4(4): R28.
doi: 10.1186/gb-2003-4-4-r28 pmid: 12702209 |
| [6] |
Antezana E, Egaña M, Blondé W, et al. The Cell Cycle Ontology: an application ontology for the representation and integrated analysis of the cell cycle process[J]. Genome Biol, 2009, 10(5): R58.
doi: 10.1186/gb-2009-10-5-r58 URL |
| [7] | Shrestha R, Arnaud E, Mauleon R, et al. Multifunctional crop trait ontology for breeders'data: field book, annotation, data discovery and semantic enrichment of the literature[J]. AoB Plants, 2010, 2010: plq008. |
| [8] |
Shrestha R, Matteis L, Skofic M, et al. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice[J]. Front Physiol, 2012, 3: 326.
doi: 10.3389/fphys.2012.00326 pmid: 22934074 |
| [9] |
Cooper L, Walls RL, Elser J, et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses[J]. Plant Cell Physiol, 2013, 54(2): e1.
doi: 10.1093/pcp/pcs163 URL |
| [10] |
Hassani-Pak K, Singh A, Brandizi M, et al. KnetMiner: a comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species[J]. Plant Biotechnol J, 2021, 19(8): 1670-1678.
doi: 10.1111/pbi.13583 pmid: 33750020 |
| [11] |
Venkatesan A, Tagny Ngompe G, Hassouni NE, et al. Agronomic Linked Data(AgroLD): a knowledge-based system to enable integrative biology in agronomy[J]. PLoS One, 2018, 13(11): e0198270.
doi: 10.1371/journal.pone.0198270 URL |
| [12] | Momtchev V, Peychev D, Primov T, et al. Expanding the pathway and interaction knowledge in linked life data[J]. Int Semant Web Chall, 2009. http://challenge.semanticweb.org/. |
| [13] | 隗玲, 胡正银, 庞弘燊, 等. 基于“主语-谓语-宾语”三元组的知识发现研究——以诱导多能干细胞领域为例[J]. 数字图书馆论坛, 2017(9): 28-34. |
| Wei L, Hu ZY, Pang HS, et al. Study on knowledge discovery based on “subject-predication-object” predications: a case study of induced pluripotent stem cells[J]. Digit Libr Forum, 2017(9): 28-34. | |
| [14] | Luciano JS, Andersson B, Batchelor C, et al. The translational medicine ontology and knowledge base: driving personalized medicine by bridging the gap between bench and bedside[J]. J Biomed Semantics, 2011, 2(Suppl 2): S1. |
| [15] | Lam HYK, Marenco L, Clark T, et al. AlzPharm: integration of neurodegeneration data using RDF[J]. BMC Bioinformatics, 2007, 8(Suppl 3): S4. |
| [16] | Pearson WR. An introduction to sequence similarity(homology)searching[J]. Curr Protoc Bioinformatics, 2013, Chapter 3: 3.1.1-3.1.8. |
| [17] |
Hu G, Kurgan L. Sequence similarity searching[J]. Curr Protoc Protein Sci, 2019, 95(1): e71.
doi: 10.1002/cpps.v95.1 URL |
| [18] |
Launay G, Simonson T. Homology modelling of protein-protein complexes: a simple method and its possibilities and limitations[J]. BMC Bioinformatics, 2008, 9: 427.
doi: 10.1186/1471-2105-9-427 pmid: 18844985 |
| [19] | Pearson WR. Selecting the right similarity-scoring matrix[J]. Curr Protoc Bioinformatics, 2013, 43: 3.5.1-3.5.9. |
| [20] |
Wu L, Zhang DF, Xue M, et al. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height[J]. J Integr Plant Biol, 2014, 56(11): 1053-1063.
doi: 10.1111/jipb.v56.11 URL |
| [21] |
Lee SM, Park CM. Regulation of reactive oxygen species generation under drought conditions in Arabidopsis[J]. Plant Signal Behav, 2012, 7(6): 599-601.
doi: 10.4161/psb.19940 URL |
| [22] |
Zheng XN, Chen B, Lu GJ, et al. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance[J]. Biochem Biophys Res Commun, 2009, 379(4): 985-989.
doi: 10.1016/j.bbrc.2008.12.163 URL |
| [23] |
Zhao X, Wu TT, Guo SX, et al. Ectopic expression of AeNAC83, a NAC transcription factor from Abelmoschus esculentus, inhibits growth and confers tolerance to salt stress in Arabidopsis[J]. Int J Mol Sci, 2022, 23(17): 10182.
doi: 10.3390/ijms231710182 URL |
| [24] |
Chen X, Lu SC, Wang YF, et al. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice[J]. Plant J, 2015, 82(2): 302-314.
doi: 10.1111/tpj.2015.82.issue-2 URL |
| [25] |
Jiang DG, Chen WT, Dong JF, et al. Overexpression of miR164b-resistant OsNAC2 improves plant architecture and grain yield in rice[J]. J Exp Bot, 2018, 69(7): 1533-1543.
doi: 10.1093/jxb/ery017 pmid: 29365136 |
| [26] |
Zhao H, Li J, Yang L, et al. An inferred functional impact map of genetic variants in rice[J]. Molecular Plant, 2021, 14: 1584-1599
doi: 10.1016/j.molp.2021.06.025 pmid: 34214659 |
| [27] |
Zhang P, Wang Y, Chachar S, et al. eRice: A refined epigenomic platform for Japonica and Indica rice[J]. Plant Biotechnology Journal, 2020, 18: 1642-1644.
doi: 10.1111/pbi.13329 pmid: 31916375 |
| [28] |
Gui S, Yang L, Li J, et al. ZEAMAP, a comprehensive database adapted to the maize multi-omics era[J]. iScience, 2020, 23(6): 101241.
doi: 10.1016/j.isci.2020.101241 URL |
| [29] |
Zhang L, Dong C, Chen Z, et al. WheatGmap: A comprehensive platform for wheat gene mapping and genomic studies[J]. Molecular Plant, 2021, 14(2): 187-190.
doi: 10.1016/j.molp.2020.11.018 pmid: 33271333 |
| [1] | Zhang Chao, Gao Shulin, Du Danni, Wu Fan, Dong Li. Isolation and Sequence Analysis of the Paeonia suffruticosa WD40 Transcription Factor Genes PsWD40-1 and PsWD40-2 [J]. Biotechnology Bulletin, 2014, 0(2): 85-90. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||