








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (11): 125-141.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0360
Previous Articles Next Articles
GU Lei( ), ZHANG Yu-lu, TANG Shang-rui, YU Hao-yue, LI Chen(
), ZHANG Yu-lu, TANG Shang-rui, YU Hao-yue, LI Chen( )
)
Received:2024-04-15
Online:2024-11-26
Published:2024-12-19
Contact:
LI Chen
E-mail:leigu@shsmu.edu.cn;cli@shsmu.edu.cn
GU Lei, ZHANG Yu-lu, TANG Shang-rui, YU Hao-yue, LI Chen. Development and Application of Mass Spectrometry-based Single-cell Proteomics Technologies[J]. Biotechnology Bulletin, 2024, 40(11): 125-141.
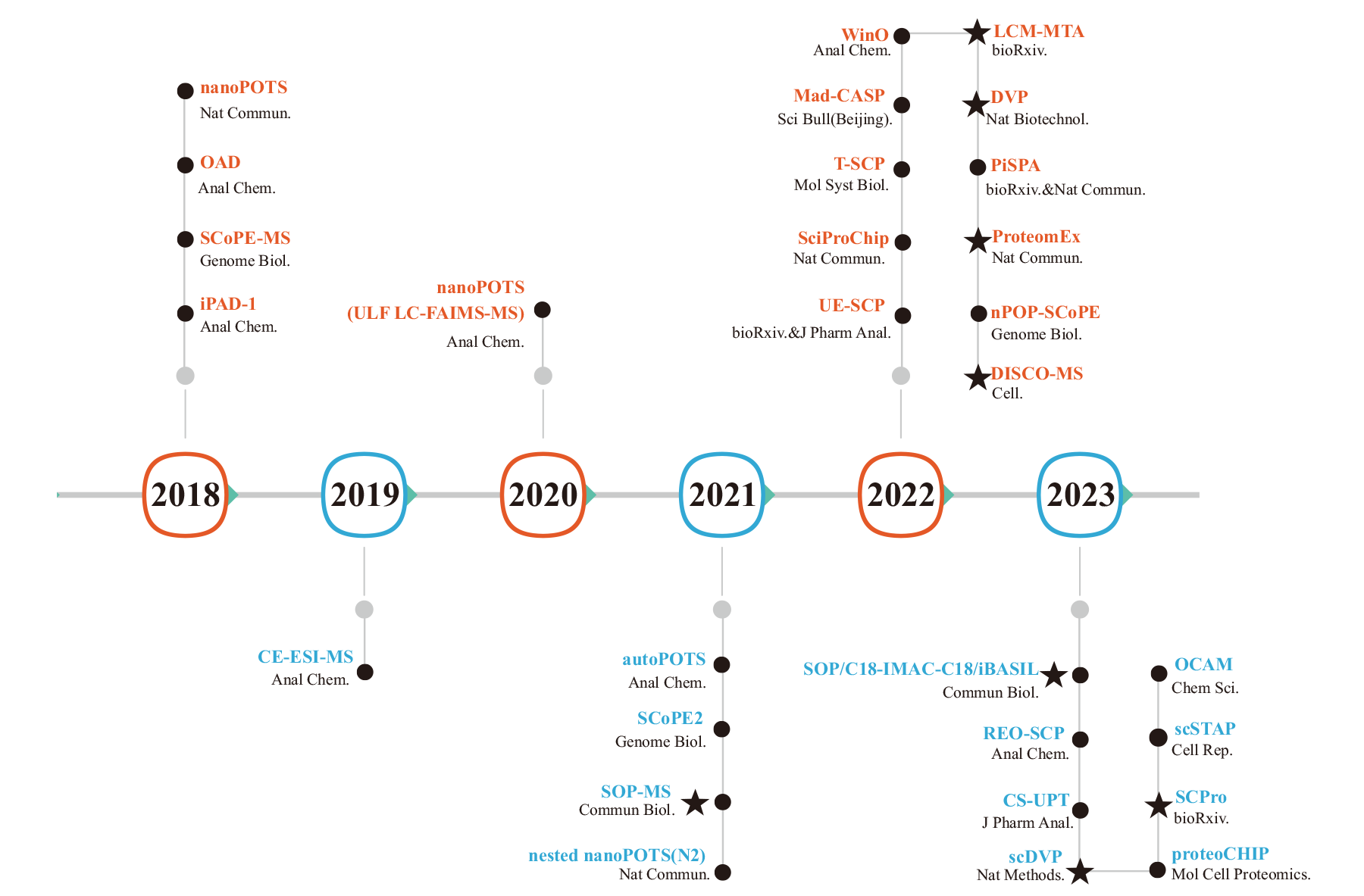
Fig. 1 Development of mass spectrometry-based single-cell proteomics technologies The figure lists representative single-cell proteomics techniques from 2018 to 2023 in chronological order; ● indicates single-cell proteomics techniques for cell suspensions, ★ indicates spatial proteomics techniques
| 单细胞蛋白质组学工具 Single-cell proteomics tools | 专用设备 Customized equipment | 标记 Label | 细胞类型 Cell types | 细胞分选分离方法 Cell isolation methods | 质谱仪 Mass spectrometers | 样品前处理通量 Pretreatment throughput(Cells per run) | 质谱检测通量 MS throughput(Cells per day) | 单个细胞中平均鉴定的蛋白质数目 Average identified protein number per cell | 单细胞蛋白质组覆盖总深度Depth of proteome coverage(n = number of cells) | 参考文献 References | 技术应用 Technical applications | 应用相关文献Related references |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| iPAD-1 | 需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Fusion Tribrid MS | 1 | 24 | 271 | 406(n=10) | [ | 图谱研究 | [ |
| OAD | 需要 | 非标记 | HeLa | 液滴微流控技术 | Orbitrap Elite MS | 1 | 4 | 51 | / | [ | 发育研究 | [ |
| nanoPOTS | 需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Fusion Lumos Tribrid MS | 27 | / | 669 | / | [ | 疾病精准分析 | [ |
| CE-ESI-MS | 需要 | 非标记 | Xenopus laevis(D11), zebrafish embryos | 毛细管电泳 | Orbitrap Q-Exactive Plus | / | / | 450-800 | / | [ | 发育研究 | [ |
| ULF LC-FAIMS-MS | 需要 | 非标记 | HeLa, U-937 | 纳升液相色谱毛细管 | Orbitrap Fusion Lumos Tribrid MS | 5 | / | 2 348 | / | [ | 癌症研究 | [ |
| nested nanoPOTS | 需要 | TMT16 | C10, RAW, SVEC | 全自动单细胞分选技术 | Orbitrap Eclipse Tribrid MS | 243 | 108 | 1 716 | 2 457(n=108) | [ | 免疫研究 | [ |
| iProChip | 需要 | 非标记 | MEC-1 | 双层聚二甲基硅氧烷(PDMS)装置 | Orbitrap Eclipse Tribrid MS | 9 | 9 | 455 | / | [ | 癌症研究 | [ |
| SciProChip | 需要 | 非标记 | PC-9 | 20 | 16 | 1 500 | 1 995(n=10) | |||||
| PiSPA | 需要 | 非标记 | A549 | 液滴微流控技术 | timsTOF Pro | 1 | / | 3 008 | 5 093(n=37) | [ | 癌症研究 | [ |
| nPOP | 需要 | TMT18 | U-937, WM989 | 全自动单细胞分选技术 | Orbitrap Q-Exactive | 2 016 | 212 | 997 | 2 844(n=1543) | [ | 癌症研究 | [ |
| proteoCHIP | 需要 | TMT16 | HeLa, HEK-293 | 全自动单细胞分选技术 | Orbitrap Exploris 480 MS | 592 | 384 | 1 940(20× carrier) | 3 674(n=276) | [ | 癌症研究 | [ |
| 1 598(no carrier) | ||||||||||||
| scSTAP | 需要 | 非标记 | 小鼠oocytes | 序控液滴微流控技术 | timsTOF Pro | / | 15-20 | 2 663 | 3 363(n=36) | [ | 发育研究 | [ |
| OCAM | 需要 | 非标记 | HeLa, 小鼠oocytes | 毛细管烷基化微反应 | Orbitrap Q-Exactive, Orbitrap Exploris 480 MS | 100 | / | 3 457 (single mouse oocyte) 1 509 (single HeLa cell) | / | [ | 发育研究 | [ |
| SCoPE | 不需要 | TMT10 | Jurkat, U-937 | 人工挑选 | LTQ Orbitrap Elite | 8 | 48 | / | 767(n=24) | [ | 图谱研究 | [ |
| SCoPE2 | 不需要 | TMT16 | Monocyte, macrophage | 流式细胞荧光分选术 | Orbitrap Q-Exactive | / | 200 | 1 000 | 3 042(n=1490) | [ | 免疫研究 | [ |
| SOP-MS | 不需要 | 非标记 | MCF10A, MCF7 | 流式细胞荧光分选术,激光捕获显微切割 | Orbitrap Q-Exactive Plus | / | / | 146 | / | [ | 癌症研究 | [ |
| SOP/C18-IMAC-C18/iBASIL | 不需要 | TMT标记 | AML | 流式细胞荧光分选术 | Orbitrap Q-Exactive Plus | / | / | 1 926 | 2 622(n=104) | [ | 免疫研究 | [ |
| autoPOTS | 不需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Exploris 480 MS | / | 10 | 301 | / | [ | 免疫研究 | [ |
| T-SCP | 不需要 | 非标记 | HeLa | 流式细胞荧光分选术 | timsTOF Pro | 308 | 41 | 2 083 | 2 501(n=231) | [ | 图谱研究 | [ |
| Mad-CASP | 不需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Eclipse Tribrid MS | / | 16 | 1 240 | / | [ | 疾病精准分析 | [ |
| WinO | 不需要 | TMT10 | RPMI8226 | 细胞分选仪 | Orbitrap Fusion Tribrid MS | / | 144 | 845 | / | [ | 免疫研究 | [ |
| UE-SCP | 不需要 | TMT6 | HeLa, HEK-293T | 全自动单细胞分选技术 | timsTOF Pro | 308 | 96 | 2 249 | 4 230(n=128) | [ | 癌症研究 | [ |
| CS-UPT | 不需要 | 非标记, TMT6, TMT8 | 小鼠MII oocytes, zygotes | 人工挑选 | timsTOF Pro | / | 18 非标记,(70, 50×car rier,100×carrier),(140, no carrier) | 2 665非标记,(2 182, 50× carrier),(2 371, 100× carrier),(1 568, no carrier) | / | [ | 发育研究 | [ |
| REO-SCP | 不需要 | 非标记 | HeLa | 全自动单细胞分选技术 | Orbitrap Exploris 480 MS | 384 | / | 1 208 | / | [ | 图谱研究 | [ |
| DVP | 不需要 | 非标记 | U2OS | 激光显微切割技术 | timsTOF Pro | / | / | 4 500-4 800(每个样品平均267个细胞核) | 5 085(8个重复) | [ | 癌症研究 | [ |
| scDVP | 不需要 | 非标记 | 小鼠肝细胞 | 激光显微切割技术 | timsTOF SCP | / | 80 | 1 700-2 700(1/3-1/2个细胞的体积) | / | [ | 图谱研究 | [ |
| LCM-MTA | 不需要 | 非标记 | 人结直肠癌癌组织与癌旁组织 | 激光显微切割技术 | timsTOF Pro | / | / | 536(15个细胞的体积) | / | [ | 癌症研究 | [ |
| ProteomEx | 不需要 | 非标记 | 小鼠脑组织 | 激光显微切割技术 | timsTOF Pro | / | / | 928(160 μm横向分辨率) | / | [ | 图谱研究 | [ |
| DISCO-MS | 不需要 | 非标记 | 小鼠脑、心脏和肺组织 | 激光显微切割技术 | timsTOF Pro | / | / | 1 400(每个ROI) | / | [ | 癌症研究 | [ |
| SCPro | 不需要 | 非标记 | HEK 293T | 激光显微切割技术 | timsTOF Pro | / | / | 800(2.4个细胞的体积) | / | [ | 疾病精准分析 | [ |
Table 1 Representative single-cell proteomics techniques and their applications
| 单细胞蛋白质组学工具 Single-cell proteomics tools | 专用设备 Customized equipment | 标记 Label | 细胞类型 Cell types | 细胞分选分离方法 Cell isolation methods | 质谱仪 Mass spectrometers | 样品前处理通量 Pretreatment throughput(Cells per run) | 质谱检测通量 MS throughput(Cells per day) | 单个细胞中平均鉴定的蛋白质数目 Average identified protein number per cell | 单细胞蛋白质组覆盖总深度Depth of proteome coverage(n = number of cells) | 参考文献 References | 技术应用 Technical applications | 应用相关文献Related references |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| iPAD-1 | 需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Fusion Tribrid MS | 1 | 24 | 271 | 406(n=10) | [ | 图谱研究 | [ |
| OAD | 需要 | 非标记 | HeLa | 液滴微流控技术 | Orbitrap Elite MS | 1 | 4 | 51 | / | [ | 发育研究 | [ |
| nanoPOTS | 需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Fusion Lumos Tribrid MS | 27 | / | 669 | / | [ | 疾病精准分析 | [ |
| CE-ESI-MS | 需要 | 非标记 | Xenopus laevis(D11), zebrafish embryos | 毛细管电泳 | Orbitrap Q-Exactive Plus | / | / | 450-800 | / | [ | 发育研究 | [ |
| ULF LC-FAIMS-MS | 需要 | 非标记 | HeLa, U-937 | 纳升液相色谱毛细管 | Orbitrap Fusion Lumos Tribrid MS | 5 | / | 2 348 | / | [ | 癌症研究 | [ |
| nested nanoPOTS | 需要 | TMT16 | C10, RAW, SVEC | 全自动单细胞分选技术 | Orbitrap Eclipse Tribrid MS | 243 | 108 | 1 716 | 2 457(n=108) | [ | 免疫研究 | [ |
| iProChip | 需要 | 非标记 | MEC-1 | 双层聚二甲基硅氧烷(PDMS)装置 | Orbitrap Eclipse Tribrid MS | 9 | 9 | 455 | / | [ | 癌症研究 | [ |
| SciProChip | 需要 | 非标记 | PC-9 | 20 | 16 | 1 500 | 1 995(n=10) | |||||
| PiSPA | 需要 | 非标记 | A549 | 液滴微流控技术 | timsTOF Pro | 1 | / | 3 008 | 5 093(n=37) | [ | 癌症研究 | [ |
| nPOP | 需要 | TMT18 | U-937, WM989 | 全自动单细胞分选技术 | Orbitrap Q-Exactive | 2 016 | 212 | 997 | 2 844(n=1543) | [ | 癌症研究 | [ |
| proteoCHIP | 需要 | TMT16 | HeLa, HEK-293 | 全自动单细胞分选技术 | Orbitrap Exploris 480 MS | 592 | 384 | 1 940(20× carrier) | 3 674(n=276) | [ | 癌症研究 | [ |
| 1 598(no carrier) | ||||||||||||
| scSTAP | 需要 | 非标记 | 小鼠oocytes | 序控液滴微流控技术 | timsTOF Pro | / | 15-20 | 2 663 | 3 363(n=36) | [ | 发育研究 | [ |
| OCAM | 需要 | 非标记 | HeLa, 小鼠oocytes | 毛细管烷基化微反应 | Orbitrap Q-Exactive, Orbitrap Exploris 480 MS | 100 | / | 3 457 (single mouse oocyte) 1 509 (single HeLa cell) | / | [ | 发育研究 | [ |
| SCoPE | 不需要 | TMT10 | Jurkat, U-937 | 人工挑选 | LTQ Orbitrap Elite | 8 | 48 | / | 767(n=24) | [ | 图谱研究 | [ |
| SCoPE2 | 不需要 | TMT16 | Monocyte, macrophage | 流式细胞荧光分选术 | Orbitrap Q-Exactive | / | 200 | 1 000 | 3 042(n=1490) | [ | 免疫研究 | [ |
| SOP-MS | 不需要 | 非标记 | MCF10A, MCF7 | 流式细胞荧光分选术,激光捕获显微切割 | Orbitrap Q-Exactive Plus | / | / | 146 | / | [ | 癌症研究 | [ |
| SOP/C18-IMAC-C18/iBASIL | 不需要 | TMT标记 | AML | 流式细胞荧光分选术 | Orbitrap Q-Exactive Plus | / | / | 1 926 | 2 622(n=104) | [ | 免疫研究 | [ |
| autoPOTS | 不需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Exploris 480 MS | / | 10 | 301 | / | [ | 免疫研究 | [ |
| T-SCP | 不需要 | 非标记 | HeLa | 流式细胞荧光分选术 | timsTOF Pro | 308 | 41 | 2 083 | 2 501(n=231) | [ | 图谱研究 | [ |
| Mad-CASP | 不需要 | 非标记 | HeLa | 流式细胞荧光分选术 | Orbitrap Eclipse Tribrid MS | / | 16 | 1 240 | / | [ | 疾病精准分析 | [ |
| WinO | 不需要 | TMT10 | RPMI8226 | 细胞分选仪 | Orbitrap Fusion Tribrid MS | / | 144 | 845 | / | [ | 免疫研究 | [ |
| UE-SCP | 不需要 | TMT6 | HeLa, HEK-293T | 全自动单细胞分选技术 | timsTOF Pro | 308 | 96 | 2 249 | 4 230(n=128) | [ | 癌症研究 | [ |
| CS-UPT | 不需要 | 非标记, TMT6, TMT8 | 小鼠MII oocytes, zygotes | 人工挑选 | timsTOF Pro | / | 18 非标记,(70, 50×car rier,100×carrier),(140, no carrier) | 2 665非标记,(2 182, 50× carrier),(2 371, 100× carrier),(1 568, no carrier) | / | [ | 发育研究 | [ |
| REO-SCP | 不需要 | 非标记 | HeLa | 全自动单细胞分选技术 | Orbitrap Exploris 480 MS | 384 | / | 1 208 | / | [ | 图谱研究 | [ |
| DVP | 不需要 | 非标记 | U2OS | 激光显微切割技术 | timsTOF Pro | / | / | 4 500-4 800(每个样品平均267个细胞核) | 5 085(8个重复) | [ | 癌症研究 | [ |
| scDVP | 不需要 | 非标记 | 小鼠肝细胞 | 激光显微切割技术 | timsTOF SCP | / | 80 | 1 700-2 700(1/3-1/2个细胞的体积) | / | [ | 图谱研究 | [ |
| LCM-MTA | 不需要 | 非标记 | 人结直肠癌癌组织与癌旁组织 | 激光显微切割技术 | timsTOF Pro | / | / | 536(15个细胞的体积) | / | [ | 癌症研究 | [ |
| ProteomEx | 不需要 | 非标记 | 小鼠脑组织 | 激光显微切割技术 | timsTOF Pro | / | / | 928(160 μm横向分辨率) | / | [ | 图谱研究 | [ |
| DISCO-MS | 不需要 | 非标记 | 小鼠脑、心脏和肺组织 | 激光显微切割技术 | timsTOF Pro | / | / | 1 400(每个ROI) | / | [ | 癌症研究 | [ |
| SCPro | 不需要 | 非标记 | HEK 293T | 激光显微切割技术 | timsTOF Pro | / | / | 800(2.4个细胞的体积) | / | [ | 疾病精准分析 | [ |
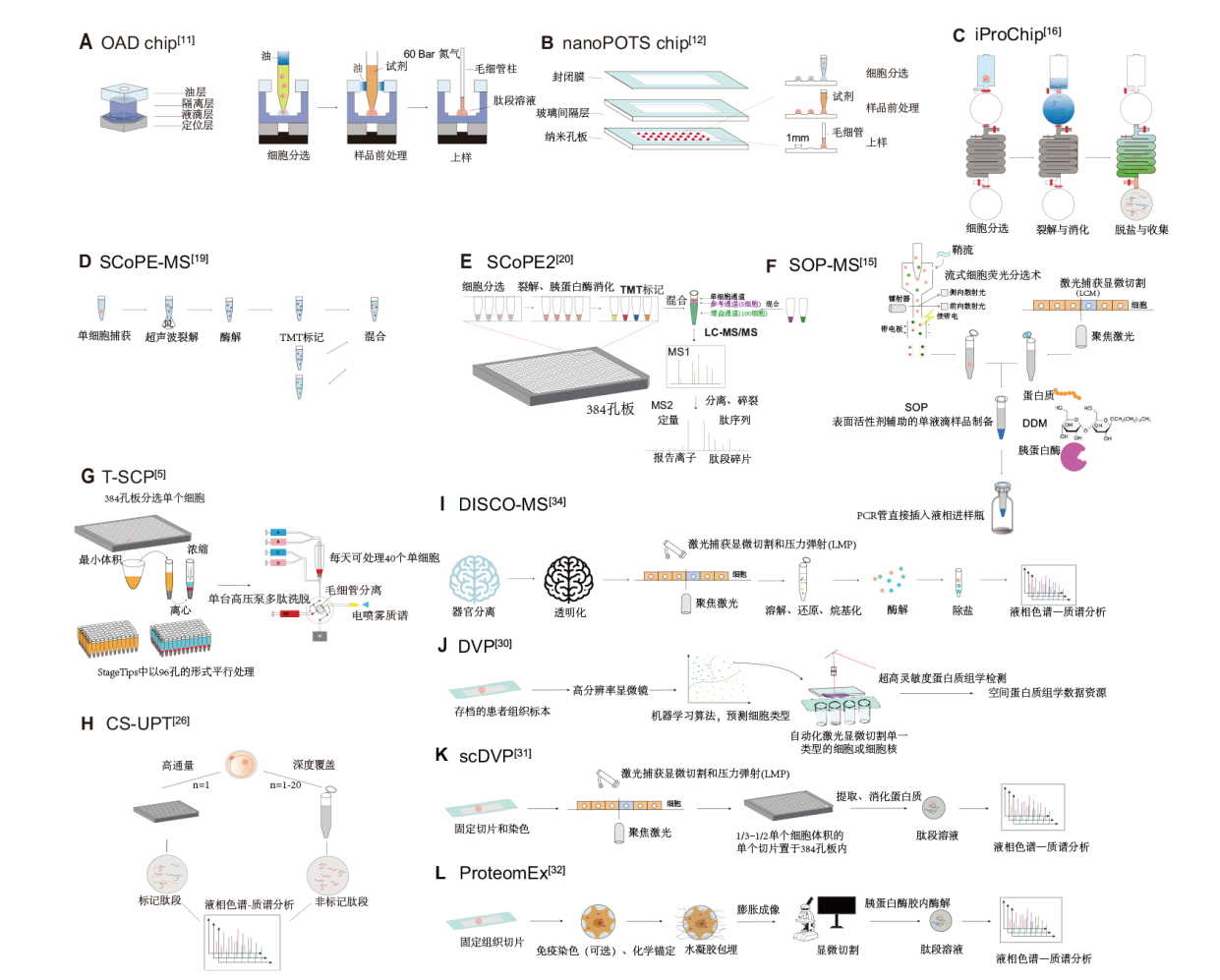
Fig. 2 Simplified schematics of representative single-cell proteomics techniques A: OAD chip is a microfluidic device used for single-cell proteomics analysis, capable of performing single-cell lysis, protein digestion, desalting, and sample collection in an integrated workflow. This chip is designed to minimize sample loss and enhance processing efficiency, especially suitable for small-volume samples. B: NanoPOTS chip is a microfluidic chip designed specifically for handling ultra-small sample volumes, suitable for proteomics sample preparation at the nanoliter and picoliter scale. This chip efficiently lyses and digests single-cell samples, significantly reducing sample loss. C: iProChip is an integrated proteomics chip used for automated operations of cell sorting, sample preparation, and mass spectrometry analysis in a microfluidic environment. The chip is designed to integrate multiple steps to improve operational efficiency and data quality. D: SCoPE-MS is a specialized technology for single-cell proteomics analysis. It uses isotope labeling to efficiently analyze multiple single-cell samples. E: SCoPE2 is an upgraded version of SCoPE-MS, featuring higher sensitivity and resolution, making it suitable for analyzing low-abundance proteins. F: SOP-MS is a mass spectrometry technique under standard operating procedures, commonly used for protein sample analysis. G: T-SCP is a high-throughput technique for single-cell proteomics, capable of processing large numbers of single-cell samples. H: iProChip is a microfluidic chip used for single-cell analysis, capable of integrating cell sorting and sample preparation in one platform. I: DISCO-MS is a technique combining tissue clearing and mass spectrometry analysis, suitable for extracting and analyzing proteins from tissue samples. J: DVP is a technique used for proteomics analysis, combining microscopy imaging with mass spectrometry. K: scDVP is the single-cell version of DVP, specifically used to analyze protein expression in single cells. L: ProteomEx is a high-resolution mass spectrometry technology used for analyzing archived tissue samples, capable of extracting single-cell information from complex tissue samples. LC-MS/MS: Liquid chromatography-tandem mass spectrometry. TMT: Tandem mass tag. SCoPE-MS: Single-cell proteomics by mass spectrometry. LMP: Laser capture microdissection and pressure catapulting. SCP: Single-cell proteomics
| 单细胞蛋白质组学数据分析软件Software for single-cell proteomics data analysis | 构建方式Construction method | 输出方式 Output method | 适配的数据采集模式 Compatible data acquisition modes | 测试数据集 Test datasets | 特点 Features | 参考文献 References |
|---|---|---|---|---|---|---|
| DO-MS | R | HTML报告 | DDA、DIA | SCoPE、SCoPE2 | 交互式可视化分析 | [ |
| SCPCompanion | C# | XML文件 | DDA | SCoPE、N2 | 推荐仪器和数据分析参数 | [ |
| SCeptre | Python | YAML文件 | DDA | AML细胞群 | 数据归一化处理 | [ |
| DART-ID | Python和R | HTML报告 | DDA | SCoPE、SCoPE2 | 对齐肽段保留时间 | [ |
| IceR | R | TSV文件 | DDA(PIP)、DIA | DeMix-Q、IonStar、HRM-DIA等 | 使用离子电流信息进行混合肽段鉴定 | [ |
| DeepSCP | Python(深度学习) | CSV文件 | DDA(TDC) | SCoPE2、N2 | 首次应用深度学习技术进行DDA分析 | [ |
Table 2 Representative software for single-cell proteomics data analysis
| 单细胞蛋白质组学数据分析软件Software for single-cell proteomics data analysis | 构建方式Construction method | 输出方式 Output method | 适配的数据采集模式 Compatible data acquisition modes | 测试数据集 Test datasets | 特点 Features | 参考文献 References |
|---|---|---|---|---|---|---|
| DO-MS | R | HTML报告 | DDA、DIA | SCoPE、SCoPE2 | 交互式可视化分析 | [ |
| SCPCompanion | C# | XML文件 | DDA | SCoPE、N2 | 推荐仪器和数据分析参数 | [ |
| SCeptre | Python | YAML文件 | DDA | AML细胞群 | 数据归一化处理 | [ |
| DART-ID | Python和R | HTML报告 | DDA | SCoPE、SCoPE2 | 对齐肽段保留时间 | [ |
| IceR | R | TSV文件 | DDA(PIP)、DIA | DeMix-Q、IonStar、HRM-DIA等 | 使用离子电流信息进行混合肽段鉴定 | [ |
| DeepSCP | Python(深度学习) | CSV文件 | DDA(TDC) | SCoPE2、N2 | 首次应用深度学习技术进行DDA分析 | [ |
| [1] |
Aslam B, Basit M, Nisar MA, et al. Proteomics: technologies and their applications[J]. J Chromatogr Sci, 2017, 55(2): 182-196.
doi: 10.1093/chromsci/bmw167 pmid: 28087761 |
| [2] |
Kelly RT. Single-cell proteomics: progress and prospects[J]. Mol Cell Proteomics, 2020, 19(11): 1739-1748.
doi: 10.1074/mcp.R120.002234 pmid: 32847821 |
| [3] |
Vistain LF, Tay S. Single-cell proteomics[J]. Trends Biochem Sci, 2021, 46(8): 661-672.
doi: 10.1016/j.tibs.2021.01.013 pmid: 33653632 |
| [4] |
Woo J, Williams SM, Markillie LM, et al. High-throughput and high-efficiency sample preparation for single-cell proteomics using a nested nanowell chip[J]. Nat Commun, 2021, 12(1): 6246.
doi: 10.1038/s41467-021-26514-2 pmid: 34716329 |
| [5] | Brunner AD, Thielert M, Vasilopoulou C, et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation[J]. Mol Syst Biol, 2022, 18(3): e10798. |
| [6] | Jiang YR, Zhu L, Cao LR, et al. Simultaneous deep transcriptome and proteome profiling in a single mouse oocyte[J]. Cell Rep, 2023, 42(11): 113455. |
| [7] |
Levy E, Slavov N. Single cell protein analysis for systems biology[J]. Essays Biochem, 2018, 62(4): 595-605.
doi: 10.1042/EBC20180014 pmid: 30072488 |
| [8] | Lundberg E, Borner GHH. Spatial proteomics: a powerful discovery tool for cell biology[J]. Nat Rev Mol Cell Biol, 2019, 20(5): 285-302. |
| [9] | Cheung TK, Lee CY, Bayer FP, et al. Defining the carrier proteome limit for single-cell proteomics[J]. Nat Meth, 2021, 18: 76-83. |
| [10] |
Shao X, Wang XT, Guan S, et al. Integrated proteome analysis device for fast single-cell protein profiling[J]. Anal Chem, 2018, 90(23): 14003-14010.
doi: 10.1021/acs.analchem.8b03692 pmid: 30375851 |
| [11] | Li ZY, Huang M, Wang XK, et al. Nanoliter-scale oil-air-droplet chip-based single cell proteomic analysis[J]. Anal Chem, 2018, 90(8): 5430-5438. |
| [12] |
Zhu Y, Piehowski PD, Zhao R, et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10-100 mammalian cells[J]. Nat Commun, 2018, 9(1): 882.
doi: 10.1038/s41467-018-03367-w pmid: 29491378 |
| [13] |
Lombard-Banek C, Moody SA, Manzini MC, et al. Microsampling capillary electrophoresis mass spectrometry enables single-cell proteomics in complex tissues: developing cell clones in live Xenopus laevis and zebrafish embryos[J]. Anal Chem, 2019, 91(7): 4797-4805.
doi: 10.1021/acs.analchem.9b00345 pmid: 30827088 |
| [14] |
Greguš M, Kostas JC, Ray S, et al. Improved sensitivity of ultralow flow LC-MS-based proteomic profiling of limited samples using monolithic capillary columns and FAIMS technology[J]. Anal Chem, 2020, 92(21): 14702-14712.
doi: 10.1021/acs.analchem.0c03262 pmid: 33054160 |
| [15] | Tsai CF, Zhang PF, Scholten D, et al. Surfactant-assisted one-pot sample preparation for label-free single-cell proteomics[J]. Commun Biol, 2021, 4(1): 265. |
| [16] |
Gebreyesus ST, Siyal AA, Kitata RB, et al. Streamlined single-cell proteomics by an integrated microfluidic chip and data-independent acquisition mass spectrometry[J]. Nat Commun, 2022, 13(1): 37.
doi: 10.1038/s41467-021-27778-4 pmid: 35013269 |
| [17] |
Wang Y, Guan ZY, Shi SW, et al. Pick-up single-cell proteomic analysis for quantifying up to 3000 proteins in a Mammalian cell[J]. Nat Commun, 2024, 15: 1279.
doi: 10.1038/s41467-024-45659-4 pmid: 38341466 |
| [18] | Tsai CF, Wang YT, Hsu CC, et al. A streamlined tandem tip-based workflow for sensitive nanoscale phosphoproteomics[J]. Commun Biol, 2023, 6: 70. |
| [19] |
Budnik B, Levy E, Harmange G, et al. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation[J]. Genome Biol, 2018, 19(1): 161.
doi: 10.1186/s13059-018-1547-5 pmid: 30343672 |
| [20] |
Specht H, Emmott E, Petelski AA, et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2[J]. Genome Biol, 2021, 22(1): 50.
doi: 10.1186/s13059-021-02267-5 pmid: 33504367 |
| [21] |
Liang YR, Acor H, McCown MA, et al. Fully automated sample processing and analysis workflow for low-input proteome profiling[J]. Anal Chem, 2021, 93(3): 1658-1666.
doi: 10.1021/acs.analchem.0c04240 pmid: 33352054 |
| [22] |
Li SQ, Su KC, Zhuang ZK, et al. A simple, rapid, and practical method for single-cell proteomics based on mass-adaptive coating of synthetic peptides[J]. Sci Bull, 2022, 67(6): 581-584.
doi: 10.1016/j.scib.2021.12.022 pmid: 36546118 |
| [23] |
Masuda T, Inamori Y, Furukawa A, et al. Water droplet-in-oil digestion method for single-cell proteomics[J]. Anal Chem, 2022, 94(29): 10329-10336.
doi: 10.1021/acs.analchem.1c05487 pmid: 35817413 |
| [24] |
Leduc A, Huffman RG, Cantlon J, et al. Exploring functional protein covariation across single cells using nPOP[J]. Genome Biol, 2022, 23(1): 261.
doi: 10.1186/s13059-022-02817-5 pmid: 36527135 |
| [25] | Gu L, Li Z, Wang Q, et al. An ultra-sensitive and easy-to-use multiplexed single-cell proteomic analysis[J]. bioRxiv, 2022. DOI: https://doi.org/10.1101/2022.01.02.474723. |
| [26] |
Gu L, Li XM, Zhu WC, et al. Ultrasensitive proteomics depicted an in-depth landscape for the very early stage of mouse maternal-to-zygotic transition[J]. J Pharm Anal, 2023, 13(8): 942-954.
doi: 10.1016/j.jpha.2023.05.003 pmid: 37719194 |
| [27] |
Matzinger M, Müller E, Dürnberger G, et al. Robust and easy-to-use one-pot workflow for label-free single-cell proteomics[J]. Anal Chem, 2023, 95(9): 4435-4445.
doi: 10.1021/acs.analchem.2c05022 pmid: 36802514 |
| [28] | Ctortecka C, Hartlmayr D, Seth A, et al. An automated nanowell-array workflow for quantitative multiplexed single-cell proteomics sample preparation at high sensitivity[J]. Mol Cell Proteomics, 2023, 22(12): 100665. |
| [29] | Gu L, Li X, Li Z, et al. Increasing the sensitivity, recovery, and integrality of spatially resolved proteomics by LCM-MTA[J]. bioRxiv, 2022. DOI: https://doi.org/10.1101/2022.08.21.504675. |
| [30] |
Mund A, Coscia F, Kriston A, et al. Deep Visual Proteomics defines single-cell identity and heterogeneity[J]. Nat Biotechnol, 2022, 40(8): 1231-1240.
doi: 10.1038/s41587-022-01302-5 pmid: 35590073 |
| [31] |
Rosenberger FA, Thielert M, Strauss MT, et al. Spatial single-cell mass spectrometry defines zonation of the hepatocyte proteome[J]. Nat Methods, 2023, 20(10): 1530-1536.
doi: 10.1038/s41592-023-02007-6 pmid: 37783884 |
| [32] |
Li L, Sun CJ, Sun YT, et al. Spatially resolved proteomics via tissue expansion[J]. Nat Commun, 2022, 13(1): 7242.
doi: 10.1038/s41467-022-34824-2 pmid: 36450705 |
| [33] | Tsai CF, Zhao R, Williams SM, et al. An improved boosting to amplify signal with isobaric labeling(iBASIL)strategy for precise quantitative single-cell proteomics[J]. Mol Cell Proteomics, 2020, 19(5): 828-838. |
| [34] |
Bhatia HS, Brunner AD, Öztürk F, et al. Spatial proteomics in three-dimensional intact specimens[J]. Cell, 2022, 185(26): 5040-5058.e19.
doi: 10.1016/j.cell.2022.11.021 pmid: 36563667 |
| [35] | Xu Y, Wang X, Li Y, et al. Multimodal single cell-resolved spatial proteomics reveals pancreatic tumor heterogeneity[J]. bioRxiv, 2023. DOI: https://doi.org/10.1101/2023.11.04.565590. |
| [36] |
Dang YJ, Zhu L, Yuan P, et al. Functional profiling of stage-specific proteome and translational transition across human pre-implantation embryo development at a single-cell resolution[J]. Cell Discov, 2023, 9(1): 10.
doi: 10.1038/s41421-022-00491-2 pmid: 36693841 |
| [37] |
He YY, Yuan HM, Liang Y, et al. On-capillary alkylation micro-reactor: a facile strategy for proteo-metabolome profiling in the same single cells[J]. Chem Sci, 2023, 14(46): 13495-13502.
doi: 10.1039/d3sc05047e pmid: 38033888 |
| [38] | Zhu WC, Ding YF, Meng J, et al. Reading and writing of mRNA m6A modification orchestrate maternal-to-zygotic transition in mice[J]. Genome Biol, 2023, 24(1): 67. |
| [39] |
Gross A, Schoendube J, Zimmermann S, et al. Technologies for single-cell isolation[J]. Int J Mol Sci, 2015, 16(8): 16897-16919.
doi: 10.3390/ijms160816897 pmid: 26213926 |
| [40] | Yokoyama WM, Christensen M, Dos Santos G, et al. Production of monoclonal antibodies[J]. Curr Protoc Immunol, 2013, 102(1): 2.5.1-2.5.29. |
| [41] | Wright G, Tucker MJ, Morton PC, et al. Micromanipulation in assisted reproduction: a review of current technology[J]. Curr Opin Obstet Gynecol, 1998, 10(3): 221-226. |
| [42] |
van den Brink SC, Sage F, Vértesy Á, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations[J]. Nat Methods, 2017, 14(10): 935-936.
doi: 10.1038/nmeth.4437 pmid: 28960196 |
| [43] |
Nakamura N, Ruebel K, Jin L, et al. Laser capture microdissection for analysis of single cells[J]. Methods Mol Med, 2007, 132: 11-18.
pmid: 17876072 |
| [44] | 宋扬, 林金明. 微流控芯片上单细胞操控与分析方法研究进展[J]. 中国科学: 化学, 2023, 53(8): 1472-1493. |
| Song Y, Lin JM. Recent advances in single-cell manipulation and analysis methods on microfluidic chips[J]. Sci Sin Chim, 2023, 53(8): 1472-1493. | |
| [45] |
Hosic S, Murthy SK, Koppes AN. Microfluidic sample preparation for single cell analysis[J]. Anal Chem, 2016, 88(1): 354-380.
doi: 10.1021/acs.analchem.5b04077 pmid: 26567589 |
| [46] |
Liu D, Sun ML, Zhang JW, et al. Single-cell droplet microfluidics for biomedical applications[J]. Analyst, 2022, 147(11): 2294-2316.
doi: 10.1039/d1an02321g pmid: 35506869 |
| [47] | Gao Y, Wu MR, Lin Y, et al. Acoustic microfluidic separation techniques and bioapplications: a review[J]. Micromachines, 2020, 11(10): 921. |
| [48] | 贺映云, 袁辉明, 梁振, 等. 单细胞蛋白质组学分析技术研究进展[J]. 分析试验室, 2022, 41(12): 1365-1378. |
| He YY, Yuan HM, Liang Z, et al. Progress in technologies for single-cell proteome analysis[J]. Chin J Anal Lab, 2022, 41(12): 1365-1378. | |
| [49] |
Shen Y, Tolicć N, Masselon C, et al. Nanoscale proteomics[J]. Anal Bioanal Chem, 2004, 378(4): 1037-1045.
pmid: 14647945 |
| [50] |
Shen YF, Zhao R, Berger SJ, et al. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics[J]. Anal Chem, 2002, 74(16): 4235-4249.
pmid: 12199598 |
| [51] |
Shen YF, Tolić N, Masselon C, et al. Ultrasensitive proteomics using high-efficiency on-line micro-SPE-nanoLC-nanoESI MS and MS/MS[J]. Anal Chem, 2004, 76(1): 144-154.
pmid: 14697044 |
| [52] |
Zhu Y, Clair G, Chrisler WB, et al. Proteomic analysis of single mammalian cells enabled by microfluidic nanodroplet sample preparation and ultrasensitive NanoLC-MS[J]. Angew Chem Int Ed, 2018, 57(38): 12370-12374.
doi: 10.1002/anie.201802843 pmid: 29797682 |
| [53] |
Cong YZ, Liang YR, Motamedchaboki K, et al. Improved single-cell proteome coverage using narrow-bore packed NanoLC columns and ultrasensitive mass spectrometry[J]. Anal Chem, 2020, 92(3): 2665-2671.
doi: 10.1021/acs.analchem.9b04631 pmid: 31913019 |
| [54] | Orsburn BC. Acetic acid is a superior acidifier for sub-nanogram and single cell proteomic studies[J]. bioRxiv, 2023. DOI: https://doi.org/10.1101/2023.08.01.551522. |
| [55] | Ye ZL, Sabatier P, Van der Hoeven L, et al. High-throughput and scalable single cell proteomics identifies over 5000 proteins per cell[J]. bioRxiv, 2023. DOI: https://doi.org/10.1101/2023.11.27.568953. |
| [56] | Bubis JA, Arrey TN, Damoc E, et al. Challenging the AstralTM mass analyzer - up to 5300 proteins per single-cell at unseen quantitative accuracy to study cellular heterogeneity[J]. bioRxiv, 2024. DOI: https://doi.org/10.1101/2024.02.01.578358 |
| [57] | Ctortecka C, Clark NM, Boyle B, et al. Automated single-cell proteomics providing sufficient proteome depth to study complex biology beyond cell type classifications[J]. bioRxiv, 2024. DOI: https://doi.org/10.1101/2024.01.20.576369. |
| [58] |
Stadlmann J, Hudecz O, Krššáková G, et al. Improved sensitivity in low-input proteomics using micropillar array-based chromatography[J]. Anal Chem, 2019, 91(22): 14203-14207.
doi: 10.1021/acs.analchem.9b02899 pmid: 31612716 |
| [59] |
Amenson-Lamar EA, Sun LL, Zhang ZB, et al. Detection of 1 zmol injection of angiotensin using capillary zone electrophoresis coupled to a Q-Exactive HF mass spectrometer with an electrokinetically pumped sheath-flow electrospray interface[J]. Talanta, 2019, 204: 70-73.
doi: S0039-9140(19)30573-9 pmid: 31357355 |
| [60] | Dou MW, Zhu Y, Liyu A, et al. Nanowell-mediated two-dimensional liquid chromatography enables deep proteome profiling of <1000 mammalian cells[J]. Chem Sci, 2018, 9(34): 6944-6951. |
| [61] |
Krieger JR, Wybenga-Groot LE, Tong JF, et al. Evosep one enables robust deep proteome coverage using tandem mass tags while significantly reducing instrument time[J]. J Proteome Res, 2019, 18(5): 2346-2353.
doi: 10.1021/acs.jproteome.9b00082 pmid: 30938160 |
| [62] |
Zheng RS, Stejskal K, Pynn C, et al. Deep single-shot NanoLC-MS proteome profiling with a 1500 bar UHPLC system, long fully porous columns, and HRAM MS[J]. J Proteome Res, 2022, 21(10): 2545-2551.
doi: 10.1021/acs.jproteome.2c00270 pmid: 36068014 |
| [63] |
Zhu Y, Zhao R, Piehowski PD, et al. Subnanogram proteomics: impact of LC column selection, MS instrumentation and data analysis strategy on proteome coverage for trace samples[J]. Int J Mass Spectrom, 2018, 427: 4-10.
doi: 10.1016/j.ijms.2017.08.016 pmid: 29576737 |
| [64] | Cox J. Prediction of peptide mass spectral libraries with machine learning[J]. Nat Biotechnol, 2023, 41(1): 33-43. |
| [65] |
Michalski A, Cox J, Mann M. More than 100, 000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS[J]. J Proteome Res, 2011, 10(4): 1785-1793.
doi: 10.1021/pr101060v pmid: 21309581 |
| [66] | Guan SH, Taylor PP, Han ZW, et al. Data dependent-independent acquisition(DDIA)proteomics[J]. J Proteome Res, 2020, 19(8): 3230-3237. |
| [67] |
Schoof EM, Furtwängler B, Üresin N, et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies[J]. Nat Commun, 2021, 12(1): 3341.
doi: 10.1038/s41467-021-23667-y pmid: 34099695 |
| [68] |
Kalxdorf M, Müller T, Stegle O, et al. IceR improves proteome coverage and data completeness in global and single-cell proteomics[J]. Nat Commun, 2021, 12(1): 4787.
doi: 10.1038/s41467-021-25077-6 pmid: 34373457 |
| [69] | Wang B, Wang Y, Chen Y, et al. DeepSCP: utilizing deep learning to boost single-cell proteome coverage[J]. Brief Bioinform, 2022, 23(4): bbac214. |
| [70] |
Huang Z, Merrihew GE, Larson EB, et al. Brain proteomic analysis implicates actin filament processes and injury response in resilience to Alzheimer’s disease[J]. Nat Commun, 2023, 14(1): 2747.
doi: 10.1038/s41467-023-38376-x pmid: 37173305 |
| [71] | Sun FY, Li HY, Sun DQ, et al. Single-cell omics: experimental workflow, data analyses and applications[J]. Sci China Life Sci, 2024. DOI: 10.1007/s11427-023-2561-0. |
| [72] |
Malioutov D, Chen TC, Airoldi E, et al. Quantifying homologous proteins and proteoforms[J]. Mol Cell Proteomics, 2019, 18(1): 162-168.
doi: 10.1074/mcp.TIR118.000947 pmid: 30282776 |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||