








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 336-346.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0008
Previous Articles Next Articles
WANG Hui1,2( ), FAN Ling-xi2, SUN Ji-lu1, WANG Yuan3, WU Ning-feng2, TIAN Jian3, GUAN Fei-fei2(
), FAN Ling-xi2, SUN Ji-lu1, WANG Yuan3, WU Ning-feng2, TIAN Jian3, GUAN Fei-fei2( )
)
Received:2025-01-04
Online:2025-07-26
Published:2025-07-22
Contact:
GUAN Fei-fei
E-mail:asherhui0120@163.com;guanfeifei@caas.cn
WANG Hui, FAN Ling-xi, SUN Ji-lu, WANG Yuan, WU Ning-feng, TIAN Jian, GUAN Fei-fei. Enhancing the Thermostability of Lysozyme RPL187 Based on Protein Intelligence Models[J]. Biotechnology Bulletin, 2025, 41(7): 336-346.

Fig. 1 Expression and activity assay of RPL187A: SDS-PAGE protein gel electrophoresis, crude enzyme solution was the heterogeneous protein produced by fragmentation after induction, and pure enzyme solution was obtained by purification. B: Oxford cup circle of inhibition experiment graph of crude enzyme solution, NC is the negative control, PC is the positive control, and CK is the blank control. C: Specific activity graph of lysin RPL187 enzyme detected by the national standard method, NC is the negative control, and PC is the positive control. ****P<0.000 1
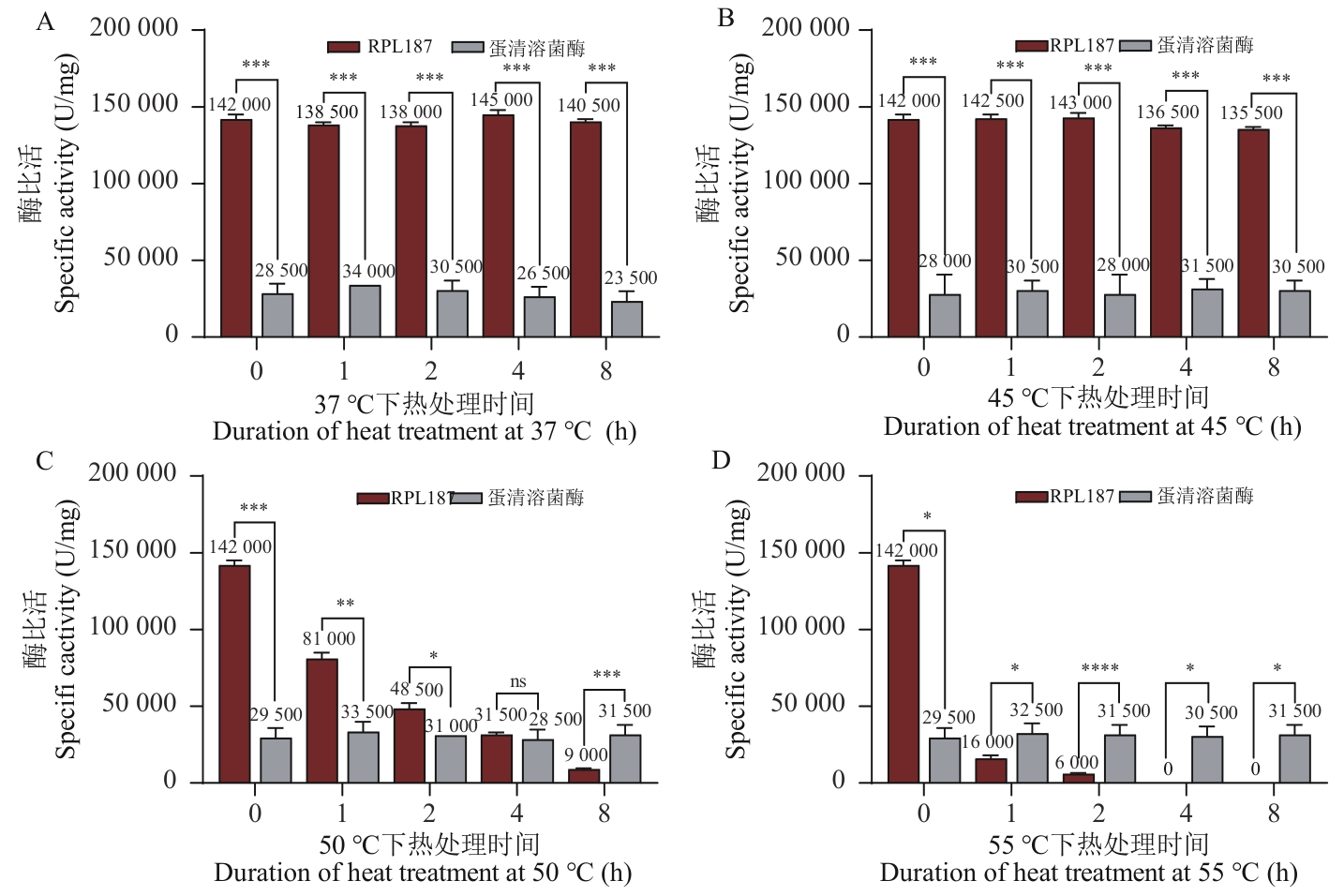
Fig. 2 Thermostability of lysozyme RPL187 by national standard methodA: Specific activity of lysozyme RPL187 and egg white lysozyme HEWL at different durations of heat treatment at 37 ℃. B: Specific activity of lysozyme RPL187 and egg white lysozyme HEWL at different durations of heat treatment at 45 ℃. C: Specific activity of lysozyme RPL187 and egg white lysozyme HEWL at different durations of heat treatment at 50 ℃. D: Specific activity of lysozyme RPL187 and egg white lysozyme at different durations of heat treatment at 55 ℃. ns: P≥0.05; *P<0.05; **P<0.01; ***P<0.001; ****P<0.000 1. The same below

Fig. 3 Expression and viability assay of the RPL187 mutantsA: SDS-PAGE protein gel electrophoresis of crude enzyme solution of RPL187 and mutants. B: Oxford cup circle of inhibition experiment graph of crude enzyme solution

Fig. 4 Thermostability of RPL187 and mutants by national standard methodA: Plots of changes in specific activity of egg white lysozyme, RPL187 and mutants RPL187-209 and RPL187-592 at different durations of heat treatment at 50 ℃. B: Plots of changes in specific activity of egg white lysozyme, RPL187 and mutants RPL187-209 and RPL187-592 at different durations of heat treatment at 55 ℃
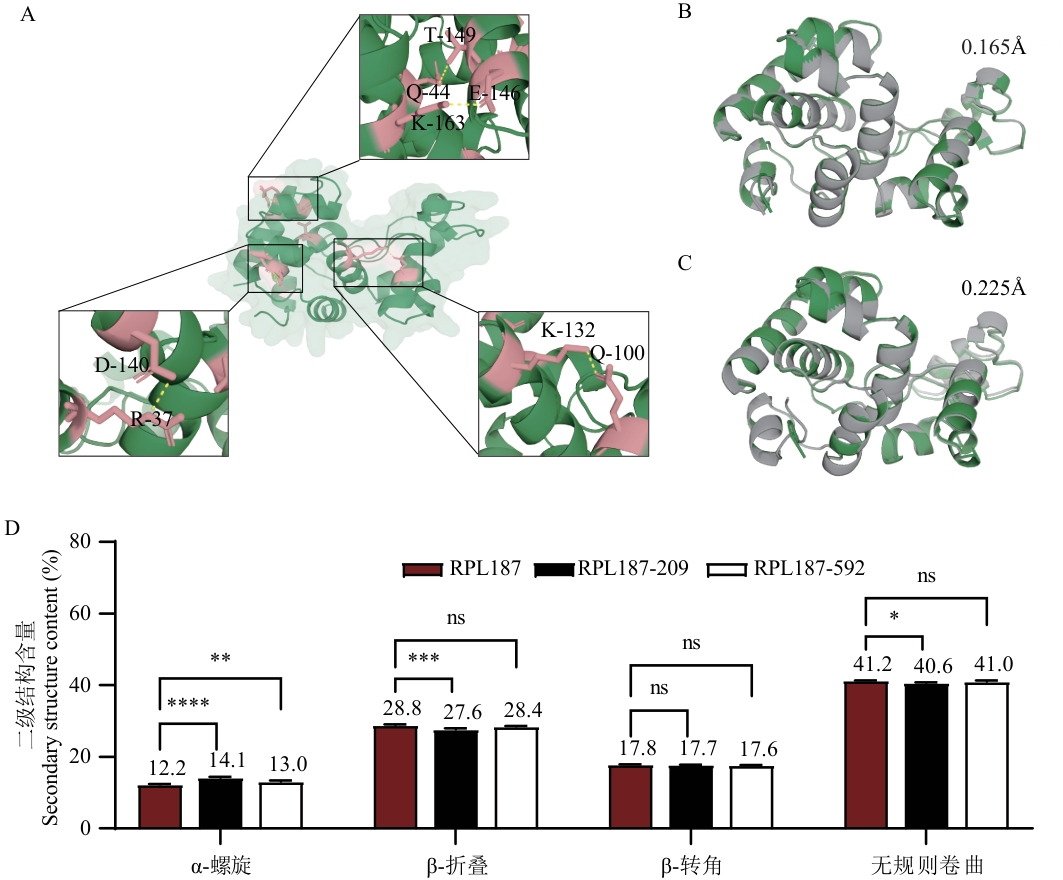
Fig. 5 Analysis diagram of the mechanism of improving thermostabilityA: Pymol visualisation showing the increased hydrogen bonding map of mutant RPL187-592 related to the wild type. B: Calculating RMSD (root mean square deviation) values of mutant RPL187-209 versus wild type RPL187 using Pymol. C: Calculating RMSD values of mutant RPL187-592 versus wild type RPL187 using Pymol. Based on the Pymol (grey) and wild-type (green) protein structure superimposed diagrams to visually demonstrate the conformational changes caused by the mutation; RMSD was used to measure the similarity between the structures of the two proteins, with smaller values indicating smaller structural differences. D: Detection of the secondary structure content changes of the wild-type RPL187 and the mutants RPL187-209 and RPL187-592 by circular dichroism (CD)
名称 Name | 模型性能 评估值 Evaluated value of model performance | Tm值 预测模型 Tm value predicting model | 最适生长温度 预测模型 Predicting model for optimal growth temperature | 大肠杆菌 表达量预测模型 Predicting model for E. coli expression | 枯草芽孢杆菌 表达量预测模型 Predicting model for B. subtilis expression | 酿酒酵母 表达量预测模型 Predicting model for S. cerevisiae expression | 总和 Sum |
|---|---|---|---|---|---|---|---|
| RPL187-592 | 0.83 | -0.69 | 2.42 | 2.08 | 1.32 | 1.60 | 6.72 |
| RPL187-319 | 0.80 | 1.26 | 1.28 | 1.18 | 0.60 | 1.79 | 6.10 |
| RPL187-496 | 0.80 | 0.49 | 0.89 | 1.98 | 0.34 | 2.18 | 5.88 |
| RPL187-622 | 0.80 | 0.13 | -0.25 | 2.31 | 0.81 | 2.63 | 5.63 |
| RPL187-168 | 0.84 | 2.15 | 1.96 | -0.65 | 1.70 | -1.34 | 3.82 |
| RPL187-857 | 0.82 | 0.95 | 1.78 | -0.87 | 1.92 | -0.38 | 3.40 |
| RPL187-649 | 0.84 | 1.69 | 1.20 | -0.65 | 1.29 | -1.02 | 2.50 |
| RPL187-954 | 0.83 | 1.09 | 0.43 | -0.54 | 0.11 | 0.27 | 1.37 |
| RPL187-209 | 0.80 | -0.96 | -0.34 | 1.71 | 0.64 | -0.01 | 1.05 |
| RPL187-963 | 0.83 | 0.22 | 0.33 | 0.18 | 0.04 | -0.27 | 0.51 |
| RPL187-700 | 0.82 | -0.93 | 0.86 | -0.60 | 0.82 | -0.12 | 0.03 |
| RPL187 | -1.00 | -1.37 | -1.25 | -1.12 | -1.58 | -1.35 | -6.67 |
Table 1 Multi-model combined screening of RPL187 mutant evaluation score sheet
名称 Name | 模型性能 评估值 Evaluated value of model performance | Tm值 预测模型 Tm value predicting model | 最适生长温度 预测模型 Predicting model for optimal growth temperature | 大肠杆菌 表达量预测模型 Predicting model for E. coli expression | 枯草芽孢杆菌 表达量预测模型 Predicting model for B. subtilis expression | 酿酒酵母 表达量预测模型 Predicting model for S. cerevisiae expression | 总和 Sum |
|---|---|---|---|---|---|---|---|
| RPL187-592 | 0.83 | -0.69 | 2.42 | 2.08 | 1.32 | 1.60 | 6.72 |
| RPL187-319 | 0.80 | 1.26 | 1.28 | 1.18 | 0.60 | 1.79 | 6.10 |
| RPL187-496 | 0.80 | 0.49 | 0.89 | 1.98 | 0.34 | 2.18 | 5.88 |
| RPL187-622 | 0.80 | 0.13 | -0.25 | 2.31 | 0.81 | 2.63 | 5.63 |
| RPL187-168 | 0.84 | 2.15 | 1.96 | -0.65 | 1.70 | -1.34 | 3.82 |
| RPL187-857 | 0.82 | 0.95 | 1.78 | -0.87 | 1.92 | -0.38 | 3.40 |
| RPL187-649 | 0.84 | 1.69 | 1.20 | -0.65 | 1.29 | -1.02 | 2.50 |
| RPL187-954 | 0.83 | 1.09 | 0.43 | -0.54 | 0.11 | 0.27 | 1.37 |
| RPL187-209 | 0.80 | -0.96 | -0.34 | 1.71 | 0.64 | -0.01 | 1.05 |
| RPL187-963 | 0.83 | 0.22 | 0.33 | 0.18 | 0.04 | -0.27 | 0.51 |
| RPL187-700 | 0.82 | -0.93 | 0.86 | -0.60 | 0.82 | -0.12 | 0.03 |
| RPL187 | -1.00 | -1.37 | -1.25 | -1.12 | -1.58 | -1.35 | -6.67 |
名称 Name | 突变数 Number of mutations | 突变氨基酸 Mutated amino acids |
|---|---|---|
| RPL187-592 | 6 | K2V__C26A__A43S__S115D__K137A__K150R |
| RPL187-319 | 8 | N3R__Q11A__C26A__Y54T__S65A__I148V__V152I__F173I |
| RPL187-496 | 4 | C26A__Y54T__K78P__C82R |
| RPL187-622 | 5 | M10L__C26A__C82R__Y89L__M111Q |
| RPL187-168 | 6 | V5I__R39A__L51S__F84Y__R109N__E146T |
| RPL187-857 | 7 | V5I__K12A__R53Q__Q56R__S65A__K137A__E146T |
| RPL187-649 | 5 | V5I__M10L__M13L__S118A__K132S |
| RPL187-954 | 7 | Q11A__R39A__A50G__L51S__S65A__F84Y__K137A |
| RPL187-209 | 4 | K78P__Q99T__K108P__K137A |
| RPL187-963 | 3 | M10L__R39A__K132S |
| RPL187-700 | 8 | K78P__A87R__Y89L__M111Q__S118A__K132S__E146T__F173I |
| RPL187 | 0 | - |
Table 2 Table of mutation site changes in RPL187 stability-modified mutants
名称 Name | 突变数 Number of mutations | 突变氨基酸 Mutated amino acids |
|---|---|---|
| RPL187-592 | 6 | K2V__C26A__A43S__S115D__K137A__K150R |
| RPL187-319 | 8 | N3R__Q11A__C26A__Y54T__S65A__I148V__V152I__F173I |
| RPL187-496 | 4 | C26A__Y54T__K78P__C82R |
| RPL187-622 | 5 | M10L__C26A__C82R__Y89L__M111Q |
| RPL187-168 | 6 | V5I__R39A__L51S__F84Y__R109N__E146T |
| RPL187-857 | 7 | V5I__K12A__R53Q__Q56R__S65A__K137A__E146T |
| RPL187-649 | 5 | V5I__M10L__M13L__S118A__K132S |
| RPL187-954 | 7 | Q11A__R39A__A50G__L51S__S65A__F84Y__K137A |
| RPL187-209 | 4 | K78P__Q99T__K108P__K137A |
| RPL187-963 | 3 | M10L__R39A__K132S |
| RPL187-700 | 8 | K78P__A87R__Y89L__M111Q__S118A__K132S__E146T__F173I |
| RPL187 | 0 | - |
名称 Name | 蛋白质熔化温度 Tm(℃) | 自由能变化 Free energy change(kcal/mol) |
|---|---|---|
| RPL187 | 52.62±0.26 | 0 |
| RPL187-209 | 54.68±0.28 | -1.57 |
| RPL187-592 | 55.03±0.28 | -0.43 |
Table 3 Changes in Tm values and free energy changes in RPL187 and mutants
名称 Name | 蛋白质熔化温度 Tm(℃) | 自由能变化 Free energy change(kcal/mol) |
|---|---|---|
| RPL187 | 52.62±0.26 | 0 |
| RPL187-209 | 54.68±0.28 | -1.57 |
| RPL187-592 | 55.03±0.28 | -0.43 |
| [1] | Wu TT, Jiang QQ, Wu D, et al. What is new in lysozyme research and its application in food industry? A review [J]. Food Chem, 2019, 274: 698-709. |
| [2] | Abdou AM, Higashiguchi S, Aboueleinin AM, et al. Antimicrobial peptides derived from hen egg lysozyme with inhibitory effect against Bacillus species [J]. Food Contr, 2007, 18(2): 173-178. |
| [3] | Abey SK, Yuana Y, Joseph PV, et al. Lysozyme association with circulating RNA, extracellular vesicles, and chronic stress [J]. BBA Clin, 2017, 7: 23-35. |
| [4] | Bahrami A, Delshadi R, Assadpour E, et al. Antimicrobial-loaded nanocarriers for food packaging applications [J]. Adv Colloid Interface Sci, 2020, 278: 102140. |
| [5] | Wu HY, Cao DN, Liu TX, et al. Purification and characterization of recombinant human lysozyme from eggs of transgenic chickens [J]. PLoS One, 2015, 10(12): e0146032. |
| [6] | Jiang MF, Hu MJ, Ren HH, et al. Molecular cloning and characterization of a new C-type lysozyme gene from yak mammary tissue [J]. Asian-Australas J Anim Sci, 2015, 28(12): 1774-1783. |
| [7] | Kim YK, Nam YK. Molecular characterization and expression pattern of c-type and g-type lysozyme isoforms in starry flounder Platichthys stellate infected with Streptococcus parauberis [J]. Fish Sci, 2015, 81: 353-363. |
| [8] | Shahmohammadi A. Lysozyme separation from chicken egg white: a review [J]. Eur Food Res Technol, 2018, 244(4): 577-593. |
| [9] | Gill AO, Holley RA. Inhibition of bacterial growth on ham and bologna by lysozyme, nisin and EDTA [J]. Food Res Int, 2000, 33(2): 83-90. |
| [10] | Ferraboschi P, Ciceri S, Grisenti P. Applications of lysozyme, an innate immune defense factor, as an alternative antibiotic [J]. Antibiotics, 2021, 10(12): 1534. |
| [11] | Ercan D, Demirci A. Recent advances for the production and recovery methods of lysozyme [J]. Crit Rev Biotechnol, 2016, 36(6): 1078-1088. |
| [12] | Salazar O, Asenjo JA. Enzymatic Lysis of microbial cells [J]. Biotechnol Lett, 2007, 29(7): 985-994. |
| [13] | Wells JE, Berry ED, Kalchayanand N, et al. Effect of lysozyme or antibiotics on faecal zoonotic pathogens in nursery pigs [J]. J Appl Microbiol, 2015, 118(6): 1489-1497. |
| [14] | Nawaz N, Wen S, Wang FH, et al. Lysozyme and its application as antibacterial agent in food industry [J]. Molecules, 2022, 27(19): 6305. |
| [15] | Elkordy AA, Forbes RT, Barry BW. Stability of crystallised and spray-dried lysozyme [J]. Int J Pharm, 2004, 278(2): 209-219. |
| [16] | 唐汉颖, 韦加娜. 新型抗菌纸的制备及其抗菌性能研究 [C]. 中国化学会, 2016:198-199. |
| Tang HY, Wei JN. Preparation of new antimicrobial paper and research on its antimicrobial properties [C]. Chinese Chemical Society, 2016: 198-199. | |
| [17] | 田健, 王平, 伍宁丰, 等. 理性设计提高蛋白质热稳定性的研究进展 [J]. 生物技术进展, 2012, 2(4): 233-239. |
| Tian J, Wang P, Wu NF, et al. Recent advances in the rational design to improve the protein thermostability [J]. Curr Biotechnol, 2012, 2(4): 233-239. | |
| [18] | Abdulkareem RA, Doekhie A, Fotaki N, et al. Thermal stabilisation of lysozyme through ensilication [J]. Molecules, 2024, 29(17): 4207. |
| [19] | Venkataramani S, Truntzer J, Coleman DR. Thermal stability of high concentration lysozyme across varying pH: a Fourier Transform Infrared study [J]. J Pharm Bioallied Sci, 2013, 5(2): 148-153. |
| [20] | Avanti C, Saluja V, van Streun ELP, et al. Stability of lysozyme in aqueous extremolyte solutions during heat shock and accelerated thermal conditions [J]. PLoS One, 2014, 9(1): e86244. |
| [21] | Steven Johnson L, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure [J]. BMC Bioinformatics, 2010, 11: 431. |
| [22] | Liu TY, Gao H, Ren XP, et al. Protein-protein interaction and site prediction using transfer learning [J]. Brief Bioinform, 2023, 24(6): bbad376. |
| [23] | Schymkowitz J, Borg J, Stricher F, et al. The FoldX web server: an online force field [J]. Nucleic Acids Res, 2005, 33(Web Server issue): W382-W388. |
| [24] | Kalayan J, Chakravorty A, Warwicker J, et al. Total free energy analysis of fully hydrated proteins [J]. Proteins, 2023, 91(1): 74-90. |
| [25] | Sun ZT, Liu Q, Qu G, et al. Utility of B-factors in protein science: interpreting rigidity, flexibility, and internal motion and engineering thermostability [J]. Chem Rev, 2019, 119(3): 1626-1665. |
| [26] | Britton KL, Baker PJ, Borges KM, et al. Insights into thermal stability from a comparison of the glutamate dehydrogenases from Pyrococcus furiosus and Thermococcus litoralis [J]. Eur J Biochem, 1995, 229(3): 688-695. |
| [27] | Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold [J]. Nature, 2021, 596(7873): 583-589. |
| [28] | Ferruz N, Schmidt S, Höcker B. ProteinTools: a toolkit to analyze protein structures [J]. Nucleic Acids Res, 2021, 49(W1): W559-W566. |
| [29] | Pace CN, Fu HL, Lee Fryar K, et al. Contribution of hydrogen bonds to protein stability [J]. Protein Sci, 2014, 23(5): 652-661. |
| [30] | Ma BH, Liu DH, Zheng MJ, et al. Development of a double-stapled peptide stabilizing both α-helix and β-sheet structures for degrading transcription factor AR-V7 [J]. JACS Au, 2024, 4(2): 816-827. |
| [31] | Burgos MI, Ochoa A, Perillo MA. β-sheet to α-helix conversion and thermal stability of β-Galactosidase encapsulated in a nanoporous silica gel [J]. Biochem Biophys Res Commun, 2019, 508(1): 270-274. |
| [32] | Yu M, Silva TC, van Opstal A, et al. The investigation of protein diffusion via H-cell microfluidics [J]. Biophys J, 2019, 116(4): 595-609. |
| [33] | Clegg JR, Peppas NA. Design of synthetic hydrogel compositions for noncovalent protein recognition [J]. ACS Appl Mater Interfaces, 2023. |
| [34] | Ho TC, Chang CC, Chan HP, et al. Hydrogels: properties and applications in biomedicine [J]. Molecules, 2022, 27(9): 2902. |
| [35] | Levashov PA, Matolygina DA, Ovchinnikova ED, et al. The bacteriolytic activity of native and covalently immobilized lysozyme against Gram-positive and Gram-negative bacteria is differentially affected by charged amino acids and glycine [J]. FEBS Open Bio, 2019, 9(3): 510-518. |
| [36] | Carmen Salinas-Garcia M, Plaza-Garrido M, Alba-Elena D, et al. Major conformational changes in the structure of lysozyme obtained from a crystal with a very low solvent content [J]. Acta Crystallogr F Struct Biol Commun, 2019, 75(Pt 11): 687-696. |
| [37] | Pal S, Mitra RK. Nonpolar hydrophobic amino acids tune the enzymatic activity of lysozyme [J]. Biophys Chem, 2022, 288: 106842. |
| [38] | Funahashi J, Takano K, Yamagata Y, et al. Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions [J]. Biochemistry, 2000, 39(47): 14448-14456. |
| [1] | YANG Yue, TAO Yan, XIE Jing, QIAN Yun-fang. Biosynthesis of Ctenopharyngodon idella C-type Lysozyme Based on Recombinant Pichia pastoris and Its Antibacterial Activity [J]. Biotechnology Bulletin, 2021, 37(12): 169-179. |
| [2] | ZHANG Chun-chen, HU Shuang-yan, RUAN Hai-hua. Expression and Renaturation of Recombinant Human Lysozyme in Escherichia coli [J]. Biotechnology Bulletin, 2020, 36(3): 153-161. |
| [3] | WANG Qiang-hou, TAO Yan, CUI Xu, YAN Qian-qian, ZHANG Ya-li. Expression of Swimming Crab C-type Lysozyme in Pichia pastoris and Its Bacteriostatic Activity [J]. Biotechnology Bulletin, 2018, 34(10): 135-142. |
| [4] | FENG Ya-dong, TAO Yan, LI Wen, CUI Xu, WANG Qiang-hou. Expression of Channel Catfish C-type Lysozyme in Pichia pastoris and Its Bacteriostatic Activity [J]. Biotechnology Bulletin, 2017, 33(7): 195-202. |
| [5] | WEN Chong-wei ZHAO Ye-qing SHI Li OUYANG Zhen. Purification of Lysozyme from Egg White by Combination of Polyethylene Glycol Precipitation and Aqueous Two-phase Extraction [J]. Biotechnology Bulletin, 2017, 33(5): 89-93. |
| [6] | WANG Yun, ZHAI Su-zhen, ZHANG Chun-lin, WANG Ji-ping. Prokaryotic Expression of i-type Lysozyme from Periplaneta americana and Preparation of Its Polyclonal Antibodies [J]. Biotechnology Bulletin, 2016, 32(1): 138-143. |
| [7] | Peng Chuanlin , Wei Chuanchuan, Wu Jianwei, Wang Yu, Xiu Jiangfan, Shang Xiaoli, Zhao Xuejun. Cloning,Expression and Sequence Analysis of Musca domestica Antifungal Peptide-1 and Musca domestica Lysozyme [J]. Biotechnology Bulletin, 2015, 31(5): 200-205. |
| [8] | Sun Lu, Liu Zhiwen, Zou Dan, Li Dan, Pan Bo, Cong Lina. Construction of Genetic Recombination for Sea Cucumber Lysozyme in Bacillus subtilis [J]. Biotechnology Bulletin, 2014, 30(6): 150-154. |
| [9] | Li Dan, Li Cheng, Sun Lu, Zou Dan, Liu Zhiwen, Cong Lina. Integrative Expression of Stichopus japonicus Lysozyme Gene in Bacillus subtilis [J]. Biotechnology Bulletin, 2014, 30(4): 139-146. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||