








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 74-81.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0095
Previous Articles Next Articles
LI Jia-yi1,2( ), LI Jin-yi1,2, BAI Xue3, BAI Ying-guo3, LIU Bo2(
), LI Jin-yi1,2, BAI Xue3, BAI Ying-guo3, LIU Bo2( ), ZHANG Zhi-wei1(
), ZHANG Zhi-wei1( )
)
Received:2025-01-22
Online:2025-08-26
Published:2025-08-14
Contact:
LIU Bo, ZHANG Zhi-wei
E-mail:ljy20220932@163.com;lfb2500@163.com;zhiweizhang2012@163.com
LI Jia-yi, LI Jin-yi, BAI Xue, BAI Ying-guo, LIU Bo, ZHANG Zhi-wei. Enhancing 5-Aminolevulinic Acid Biosynthesis through Tandem Rare Codon-mediated Attenuation of HemB Expression[J]. Biotechnology Bulletin, 2025, 41(8): 74-81.
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| hemB-LF1 | TGATGGGTCTTGTTGTTGGCGTACCAGAACTGATCGACGCTCT | 扩增hemB左侧同源臂 |
| hemB-LR24 | TAGCCTTAGGGATAGGGACCTTAGCATTGCTTGTAACCTTTGCTCTCT | 扩增hemB左侧同源臂 |
| hemB-RF24 | CTAAGGTCCCTATCCCTAAGGCTAAGCACTTCTTCTGATTACTCCCAC | 扩增在hemB起始密码子ATG后添加24 bp DNA序列的hemB左侧同源臂 |
| hemB-RR1 | GTCCTCTGTATGTTGATAAACGCCCGGCATGGAG | 扩增在hemB起始密码子ATG后添加24 bp DNA序列的hemB左侧同源臂 |
| hemB-LF2 | GCGTTTATCAACATACAGAGGACTCGCTGCTGCGTGCCGCCCA | 扩增hemB右侧同源臂 |
| hemB-RR2 | AGTATCTTCCTGGCATCTTCCAGTCGCAGGATCTTGCTGGTAG | 扩增hemB右侧同源臂 |
| hemB-LR18 | AGTGCTTAGGGATAGGGACCTTAGCATTGCTTGTAACCTTTGCTCTCT | 扩增在hemB起始密码子ATG后添加18 bp DNA序列的hemB左侧同源臂 |
| hemB-RF18 | CTAAGGTCCCTATCCCTAAGCACTTCTTCTGATTACTCCCAC | 扩增在hemB起始密码子ATG后添加18 bp DNA序列的hemB左侧同源臂 |
| hemB-LR9 | ATCAGAAGAAGTGCTGGACCTTAGCATTGCTTGTAACCTTTGCTCTCT | 扩增在hemB起始密码子ATG后添加9 bp DNA序列的hemB左侧同源臂 |
| hemB-RF9 | CTAAGGTCCAGCACTTCTTCTGATTACTCCCAC | 扩增在hemB起始密码子ATG后添加9 bp DNA序列的hemB左侧同源臂 |
| pA15-F | CTGGAAGATGCCAGGAAGATACT | 扩增大肠杆菌复制元件和gDNA |
| hemB/gDNA-R | CTCTCATCCGCCAAAACAGCCAGCAGCGAATCTTCAGTGTGCTGAATCTACAACAGTAGAAATTCGGATCC | 扩增大肠杆菌复制元件和gDNA |
Table 1 Primers used in this study
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| hemB-LF1 | TGATGGGTCTTGTTGTTGGCGTACCAGAACTGATCGACGCTCT | 扩增hemB左侧同源臂 |
| hemB-LR24 | TAGCCTTAGGGATAGGGACCTTAGCATTGCTTGTAACCTTTGCTCTCT | 扩增hemB左侧同源臂 |
| hemB-RF24 | CTAAGGTCCCTATCCCTAAGGCTAAGCACTTCTTCTGATTACTCCCAC | 扩增在hemB起始密码子ATG后添加24 bp DNA序列的hemB左侧同源臂 |
| hemB-RR1 | GTCCTCTGTATGTTGATAAACGCCCGGCATGGAG | 扩增在hemB起始密码子ATG后添加24 bp DNA序列的hemB左侧同源臂 |
| hemB-LF2 | GCGTTTATCAACATACAGAGGACTCGCTGCTGCGTGCCGCCCA | 扩增hemB右侧同源臂 |
| hemB-RR2 | AGTATCTTCCTGGCATCTTCCAGTCGCAGGATCTTGCTGGTAG | 扩增hemB右侧同源臂 |
| hemB-LR18 | AGTGCTTAGGGATAGGGACCTTAGCATTGCTTGTAACCTTTGCTCTCT | 扩增在hemB起始密码子ATG后添加18 bp DNA序列的hemB左侧同源臂 |
| hemB-RF18 | CTAAGGTCCCTATCCCTAAGCACTTCTTCTGATTACTCCCAC | 扩增在hemB起始密码子ATG后添加18 bp DNA序列的hemB左侧同源臂 |
| hemB-LR9 | ATCAGAAGAAGTGCTGGACCTTAGCATTGCTTGTAACCTTTGCTCTCT | 扩增在hemB起始密码子ATG后添加9 bp DNA序列的hemB左侧同源臂 |
| hemB-RF9 | CTAAGGTCCAGCACTTCTTCTGATTACTCCCAC | 扩增在hemB起始密码子ATG后添加9 bp DNA序列的hemB左侧同源臂 |
| pA15-F | CTGGAAGATGCCAGGAAGATACT | 扩增大肠杆菌复制元件和gDNA |
| hemB/gDNA-R | CTCTCATCCGCCAAAACAGCCAGCAGCGAATCTTCAGTGTGCTGAATCTACAACAGTAGAAATTCGGATCC | 扩增大肠杆菌复制元件和gDNA |

Fig. 2 Schematic diagram of the strategy after inserting rare codons into starting codon hemBA: DNA sequence of hemB and its adjacent DNA sequence. B: DNA sequence of hemB and its adjacent DNA sequence in the gene-editing plasmid. In figure A, the ATG highlighted with a yellow background indicates the start codon of hemB, while the TTTA marked with a blue background indicates the PAM motif recognized by Cas12a. The gray-highlighted region in panel A corresponds to the gDNA sequence. In figure B, the gray-highlighted region shows mutations introduced into the gDNA based on the synonymous codon principle to avoid cleavage by Cas12a
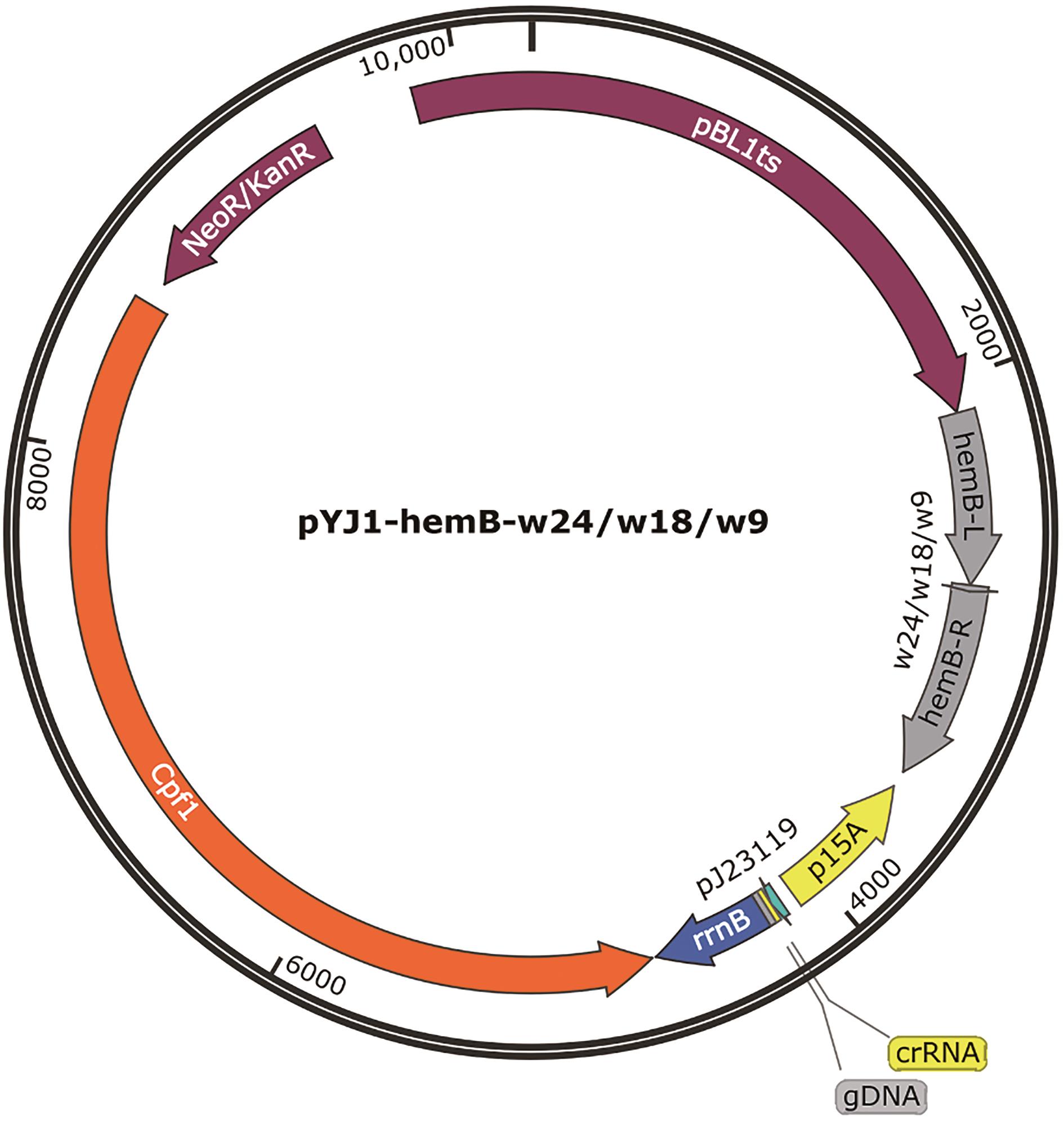
Fig. 3 Plasmid for gene editing and attenuating expression of HemBhemB-L and hemB-R as the left and right homologous sequence of hemB, respectively. The DNA sequences between hemB-L and hemB-R used for attenuating expressions of HemB as follows, w9: 5′-CTAAGGTCC-3′, w18: 5′- CTAAGGTCCCTATCCCTA-3′, w24: 5′-CTAAGGTCCCTATCCCTAAGGCTA-3′, and gDNA: 5′-TCAGCACACTGAAGATTCGCTGCTG-3′
| [1] | Tang HT, Lin SM, Deng JL, et al. Engineering yeast for the de novo synthesis of jasmonates [J]. Nat Synth, 2023, 3(2): 224-235. |
| [2] | Yang D, Park SY, Park YS, et al. Metabolic engineering of Escherichia coli for natural product biosynthesis [J]. Trends Biotechnol, 2020, 38(7): 745-765. |
| [3] | Wang K, Song XT, Cui BY, et al. Metabolic engineering of Escherichia coli for efficient production of ectoine [J]. J Agric Food Chem, 2025, 73(1): 646-654. |
| [4] | Chen JZ, Wang Y, Guo X, et al. Efficient bioproduction of 5-aminolevulinic acid, a promising biostimulant and nutrient, from renewable bioresources by engineered Corynebacterium glutamicum [J]. Biotechnol Biofuels, 2020, 13: 41. |
| [5] | Das S, Sharma K, Sharmmah D, et al. Metabolic rewiring of microbial cell factories for improved production of succinic acid [J]. Biotechnol Sustain Mater, 2024, 1(1): 15. |
| [6] | Nevoigt E, Kohnke J, Fischer CR, et al. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae [J]. Appl Environ Microbiol, 2006, 72(8): 5266-5273. |
| [7] | Lata H, Sharma A, Chadha S, et al. RNA interference (RNAi) mechanism and application in vegetable crops [J]. J Hortic Sci Biotechnol, 2022, 97(2): 160-170. |
| [8] | Tang Q, Khvorova A. RNAi-based drug design: considerations and future directions [J]. Nat Rev Drug Discov, 2024, 23(5): 341-364. |
| [9] | Zheng Y, Shen W, Zhang J, et al. CRISPR interference-based specific and efficient gene inactivation in the brain [J]. Nat Neurosci, 2018, 21(3): 447-454. |
| [10] | Izert MA, Klimecka MM, Górna MW. Applications of bacterial degrons and degraders - toward targeted protein degradation in bacteria [J]. Front Mol Biosci, 2021, 8: 669762. |
| [11] | Sabi R, Tuller T. Modelling the efficiency of codon-tRNA interactions based on codon usage bias [J]. DNA Res, 2014, 21(5): 511-526. |
| [12] | Zhang SY, Lin R, Cui LY, et al. Alter codon bias of the P. pastoris genome to overcome a bottleneck in codonoptimization strategy development and improve protein expression [J]. Microbiol Res, 2024, 282: 127629. |
| [13] | Fu HG, Liang YB, Zhong XQ, et al. Codonoptimization with deep learning to enhance protein expression [J]. Sci Rep, 2020, 10(1): 17617. |
| [14] | Xu YC, Liu KS, Han Y, et al. Codon usage bias regulates gene expression and protein conformation in yeast expression system P. pastoris [J]. Microb Cell Fact, 2021, 20(1): 91. |
| [15] | Zhang XY, Xi ZW, Zhao HT, et al. Efficient heterologous expression of bovine lactoferrin in Pichia pastoris and characterization of antibacterial activity [J]. Syst Microbiol Biomanuf, 2025, 5(1): 237-248. |
| [16] | Zhang JL, Kang Z, Chen J, et al. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli [J]. Sci Rep, 2015, 5: 8584. |
| [17] | Jiang MR, Hong KQ, Mao YF, et al. Natural 5-aminolevulinic acid: sources, biosynthesis, detection and applications [J]. Front Bioeng Biotechnol, 2022, 10: 841443. |
| [18] | Wu Y, Liao WB, Dawuda MM, et al. 5-Aminolevulinic acid (ALA) biosynthetic and metabolic pathways and its role in higher plants: a review [J]. Plant Growth Regul, 2019, 87(2): 357-374. |
| [19] | Wang JW, He YM, Wang GZ, et al. Exogenous 5-aminolevulinic acid promotes carotenoid accumulation in tomato fruits by regulating ethylene biosynthesis and signaling [J]. Physiol Plant, 2024, 176(6): e14648. |
| [20] | Casas A. Clinical uses of 5-aminolaevulinic acid in photodynamic treatment and photodetection of cancer: a review [J]. Cancer Lett, 2020, 490: 165-173. |
| [21] | Yadav V, Mai Y, McCoubrey LE, et al. 5-aminolevulinic acid as a novel therapeutic for inflammatory bowel disease [J]. Biomedicines, 2021, 9(5): 578. |
| [22] | Zou YL, Chen T, Feng LL, et al. Enhancement of 5-aminolevulinic acid production by metabolic engineering of the Glycine biosynthesis pathway in Corynebacterium glutamicum [J]. Biotechnol Lett, 2017, 39(9): 1369-1374. |
| [23] | Ding WW, Weng HJ, Du GC, et al. 5-Aminolevulinic acid production from inexpensive glucose by engineering the C4 pathway in Escherichia coli [J]. J Ind Microbiol Biotechnol, 2017, 44(8): 1127-1135. |
| [24] | Ramzi AB, Hyeon JE, Kim SW, et al. 5-Aminolevulinic acid production in engineered Corynebacterium glutamicum via C5 biosynthesis pathway [J]. Enzyme Microb Technol, 2015, 81: 1-7. |
| [25] | Luo ZS, Pan F, Zhu YF, et al. Synergistic improvement of 5-aminolevulinic acid production with synthetic scaffolds and system pathway engineering [J]. ACS Synth Biol, 2022, 11(8): 2766-2778. |
| [26] | Erskine PT, Senior N, Maignan S, et al. Crystallization of 5-aminolaevulinic acid dehydratase from Escherichia coli and Saccharomyces cerevisiae and preliminary X-ray characterization of the crystals [J]. Protein Sci, 1997, 6(8): 1774-1776. |
| [27] | Miscevic D, Mao JY, Kefale T, et al. Strain engineering for high-level 5-aminolevulinic acid production in Escherichia coli [J]. Biotechnol Bioeng, 2021, 118(1): 30-42. |
| [28] | Replogle JM, Bonnar JL, Pogson AN, et al. Maximizing CRISPRi efficacy and accessibility with dual-sgRNA libraries and optimal effectors [J]. eLife, 2022, 11: e81856. |
| [29] | Pu W, Chen JZ, Zhou YY, et al. Systems metabolic engineering of Escherichia coli for hyper-production of 5-aminolevulinic acid [J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 31. |
| [30] | Zhang J, Yang FY, Yang YP, et al. Optimizing a CRISPR-Cpf1-based genome engineering system for Corynebacterium glutamicum [J]. Microb Cell Fact, 2019, 18(1): 60. |
| [31] | Cardiff RAL, Carothers JM, Zalatan JG, et al. Systems-level modeling for CRISPR-based metabolic engineering [J]. ACS Synth Biol, 2024, 13(9): 2643-2652. |
| [32] | Cui SX, Lv XQ, Xu XH, et al. Multilayer genetic circuits for dynamic regulation of metabolic pathways [J]. ACS Synth Biol, 2021, 10(7): 1587-1597. |
| [33] | Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system [J]. J Mol Biol, 1981, 151(3): 389-409. |
| [34] | Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression [J]. Trends Biotechnol, 2004, 22(7): 346-353. |
| [35] | Kim S, Lee SB. Rare codon clusters at 5'-end influence heterologous expression of archaeal gene in Escherichia coli [J]. Protein Expr Purif, 2006, 50(1): 49-57. |
| [1] | WU Shu-ning, SU Yong-ping, LI Dong-xue, BAI Ying-guo, LIU Bo, ZHANG Zhi-wei. Design and Application of a Cumate-inducible Promoter for Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2024, 40(7): 108-116. |
| [2] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [3] | LIU Jia-hui, LIU Ye, HUA Er-bing, WANG Meng. PAM Extension of Cytosine Base Editing Tool in Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2023, 39(9): 49-57. |
| [4] | NIE Zhi-hua, ZHU Lei-lei. Effects of Biotin on L-Glutamate Efflux Mediated by MscCG in Fermentation Process [J]. Biotechnology Bulletin, 2020, 36(10): 150-155. |
| [5] | RAO De-ming, ZHANG Liang-cheng, CHEN Jiu-zhou, SUN De-hu, SUN Cun-min, ZHENG Xiao-mei, ZHENG Ping, DIAO Ai-po. Construction of 5-aminolevulinic Acid Synthesis Pathway and Optimization of Fermentation by Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2017, 33(1): 148-156. |
| [6] | Wang Shishan, Chen Yanke, Yang Jun. The Gene Sequence Optimization of Membrane Protein in Prokaryotic Expression System [J]. Biotechnology Bulletin, 2015, 31(12): 50-55. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||