








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 82-94.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0500
Previous Articles Next Articles
TIAN Yun1( ), KONG Chen1, YANG Chong2, LIU Tong-gao2, GAO Hong-rui1, LI Jia-hui1, MA Yun1, CAI Bei1(
), KONG Chen1, YANG Chong2, LIU Tong-gao2, GAO Hong-rui1, LI Jia-hui1, MA Yun1, CAI Bei1( )
)
Received:2025-05-14
Online:2025-12-26
Published:2026-01-06
Contact:
CAI Bei
E-mail:2294928895@qq.com;caibei1115@163.com
TIAN Yun, KONG Chen, YANG Chong, LIU Tong-gao, GAO Hong-rui, LI Jia-hui, MA Yun, CAI Bei. CRISPR-based High-throughput Screening Technology and Its Applications in Livestock Research[J]. Biotechnology Bulletin, 2025, 41(12): 82-94.
特征 Feature | 全基因组sgRNA文库 Genome-wide sgRNA library | 单一基因的sgRNA Single gene sgRNA |
|---|---|---|
| 目标范围 | 全基因组范围(包含数万的基因) | 单一或少数基因 |
| sgRNA数量级 | 数万或数十万条sgRNA | 4-10条sgRNA |
| 设计深度 | 每个基因4-6条sgRNA | 每个基因可设计数10条sgRNA |
| 覆盖范围 | 广度优先(尽可能的覆盖多的基因) | 深度优先(聚焦特定基因) |
| 应用场景 | 未知基因的挖掘 | 基因功能的精细验证 |
Table 1 Comparison of genome-wide sgRNA library and single gene sgRNA design
特征 Feature | 全基因组sgRNA文库 Genome-wide sgRNA library | 单一基因的sgRNA Single gene sgRNA |
|---|---|---|
| 目标范围 | 全基因组范围(包含数万的基因) | 单一或少数基因 |
| sgRNA数量级 | 数万或数十万条sgRNA | 4-10条sgRNA |
| 设计深度 | 每个基因4-6条sgRNA | 每个基因可设计数10条sgRNA |
| 覆盖范围 | 广度优先(尽可能的覆盖多的基因) | 深度优先(聚焦特定基因) |
| 应用场景 | 未知基因的挖掘 | 基因功能的精细验证 |
sgRNA文库类型 Type of sgRNA library | sgRNA位点要求 Requirements of sgRNA site | 位点要求的原因及目的 Reason and purpose of site requirements |
|---|---|---|
| 全基因组文库 | 一般位于编码序列的前25% | 确保蛋白功能的完全丧失 |
| 激活文库 | TSS上游200 bp左右 | 确保激活结构域能够靶向TSS位点,以实现基因表达的增强 |
| 干扰文库 | TSS下游+25-75 nts左右 | 确保转录抑制结构域能够靶向TSS位点,以实现沉默基因 |
| 敲除文库 | 选择编码区前50%(靠近ATG),避开内含子剪切位点 | 确保蛋白功能的丧失,避免截留的蛋白残基仍存有功能 |
| STOP文库 | TSS下游100 bp左右 | 确保终止密码子的提前引入,以丧失沉默基因。 |
Table 2 sgRNA site design requirements
sgRNA文库类型 Type of sgRNA library | sgRNA位点要求 Requirements of sgRNA site | 位点要求的原因及目的 Reason and purpose of site requirements |
|---|---|---|
| 全基因组文库 | 一般位于编码序列的前25% | 确保蛋白功能的完全丧失 |
| 激活文库 | TSS上游200 bp左右 | 确保激活结构域能够靶向TSS位点,以实现基因表达的增强 |
| 干扰文库 | TSS下游+25-75 nts左右 | 确保转录抑制结构域能够靶向TSS位点,以实现沉默基因 |
| 敲除文库 | 选择编码区前50%(靠近ATG),避开内含子剪切位点 | 确保蛋白功能的丧失,避免截留的蛋白残基仍存有功能 |
| STOP文库 | TSS下游100 bp左右 | 确保终止密码子的提前引入,以丧失沉默基因。 |
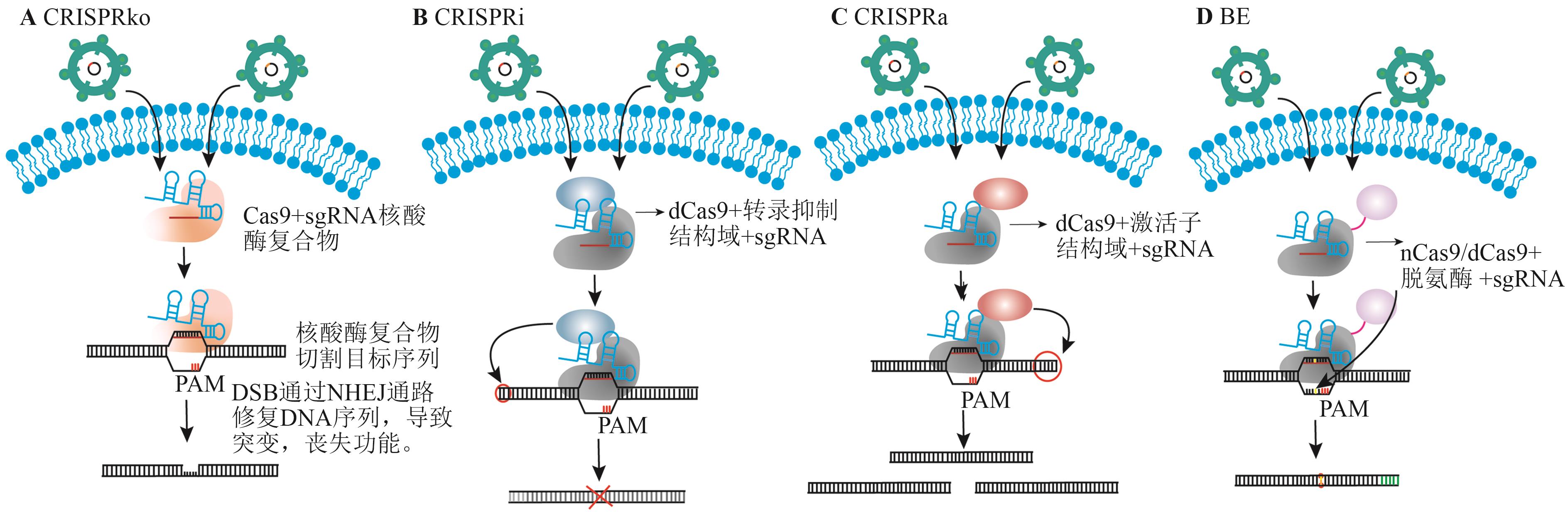
Fig. 3 Schematic diagrams of different types of CRISPR screeening toolsA: CRISPRko screening diagram. B: CRISPRi screening diagram. C: CRISPRa screening diagram. D: BE screening diagram
筛选工具 Screening tool | 扰动方式 Disturbed mode | 优点 Merit | 局限性 Limitation |
|---|---|---|---|
| RNAi | 通过降解mRNA抑制基因表达 | 简化了筛选方法,降低筛选文库的构建成本 | 无法针对基因组基因进行筛选;基因沉默不完全,只能在细胞质中行使功能 |
| cDNA/ORF | 提高目标蛋白的表达 | 第一个功能获得型筛选方法,是发现冗余基因功能的方法 | 文库构建困难,并且成本高;某些基因过表达后会导致细胞死亡 |
| CRISPRko | 利用gRNA引导Cas9酶切割目标DNA序列,通过细胞的DNA修复机制导致目标基因敲除 | 可以最大程度地避免基因的假沉默;可实现非编码基因的筛选 | 存在脱靶效应;断裂DNA双链引起的DNA毒性;无法研究基因恢复后的情况 |
| CRISPRi | 通过阻断转录起始实现基因沉默 | 不引起DNA双链断裂,造成的损伤是可逆的,脱靶效应小,特异性强 | 对基因结构有依赖性 |
| CRISPRa | 激活转录因子,增强基因转录活性 | 不引起DNA双链断裂;造成的损伤是可逆的 | 不能针对特定转录本激活;激活效率受限 |
| BE | 将脱氨酶与dCas9结合,实现碱基的转换,从而实现起始密码子和终止密码子的沉默和提前终止,以实现基因的沉默 | 不引起DNA双链断裂的情况下,实现特定碱基的转换 | 编辑范围有限;在编辑窗口中发生旁观者编辑 |
Table 3 Screening tool
筛选工具 Screening tool | 扰动方式 Disturbed mode | 优点 Merit | 局限性 Limitation |
|---|---|---|---|
| RNAi | 通过降解mRNA抑制基因表达 | 简化了筛选方法,降低筛选文库的构建成本 | 无法针对基因组基因进行筛选;基因沉默不完全,只能在细胞质中行使功能 |
| cDNA/ORF | 提高目标蛋白的表达 | 第一个功能获得型筛选方法,是发现冗余基因功能的方法 | 文库构建困难,并且成本高;某些基因过表达后会导致细胞死亡 |
| CRISPRko | 利用gRNA引导Cas9酶切割目标DNA序列,通过细胞的DNA修复机制导致目标基因敲除 | 可以最大程度地避免基因的假沉默;可实现非编码基因的筛选 | 存在脱靶效应;断裂DNA双链引起的DNA毒性;无法研究基因恢复后的情况 |
| CRISPRi | 通过阻断转录起始实现基因沉默 | 不引起DNA双链断裂,造成的损伤是可逆的,脱靶效应小,特异性强 | 对基因结构有依赖性 |
| CRISPRa | 激活转录因子,增强基因转录活性 | 不引起DNA双链断裂;造成的损伤是可逆的 | 不能针对特定转录本激活;激活效率受限 |
| BE | 将脱氨酶与dCas9结合,实现碱基的转换,从而实现起始密码子和终止密码子的沉默和提前终止,以实现基因的沉默 | 不引起DNA双链断裂的情况下,实现特定碱基的转换 | 编辑范围有限;在编辑窗口中发生旁观者编辑 |
| [1] | 王文月, 米晓钰, 孙康泰, 等. 畜禽重要性状遗传调控机制与分子设计育种 [J]. 中国农业科技导报, 2022, 24(12): 39-47. |
| Wang WY, Mi XY, Sun KT, et al. Genetic regulation mechanisms of important traits and molecular design breeding in livestock and poultry [J]. J Agric Sci Technol, 2022, 24(12): 39-47. | |
| [2] | Martiny JBH, Jones SE, Lennon JT, et al. Microbiomes in light of traits: a phylogenetic perspective [J]. Science, 2015, 350(6261): aac9323. |
| [3] | Yang CT, Lei Y, Ren TL, et al. The current situation and development prospect of whole-genome screening [J]. Int J Mol Sci, 2024, 25(1): 658. |
| [4] | Yang WN, Feng H, Zhang XH, et al. Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives [J]. Mol Plant, 2020, 13(2): 187-214. |
| [5] | Hu SJ, Gan ML, Wei ZA, et al. Identification of host factors for livestock and poultry viruses: genome-wide screening technology based on the CRISPR system [J]. Front Microbiol, 2024, 15: 1498641. |
| [6] | Unniyampurath U, Pilankatta R, Krishnan MN. RNA interference in the age of CRISPR: will CRISPR interfere with RNAi? [J]. Int J Mol Sci, 2016, 17(3): 291. |
| [7] | Vercauteren S, Fiesack S, Maroc L, et al. The rise and future of CRISPR-based approaches for high-throughput genomics [J]. FEMS Microbiol Rev, 2024, 48(5): fuae020. |
| [8] | St Johnston D. The art and design of genetic screens: Drosophila melanogaster [J]. Nat Rev Genet, 2002, 3(3): 176-188. |
| [9] | Jorgensen EM, Mango SE. The art and design of genetic screens: Caenorhabditis elegans [J]. Nat Rev Genet, 2002, 3(5): 356-369. |
| [10] | Forsburg SL. The art and design of genetic screens: yeast [J]. Nat Rev Genet, 2001, 2(9): 659-668. |
| [11] | Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9 [J]. Nat Rev Genet, 2015, 16(5): 299-311. |
| [12] | Josi C, Bürki S, Vidal S, et al. Large-scale analysis of the Mycoplasma bovis genome identified non-essential, adhesion- and virulence-related genes [J]. Front Microbiol, 2019, 10: 2085. |
| [13] | Hai T, Cao C, Shang H, et al. Pilot study of large-scale production of mutant pigs by ENU mutagenesis [J]. eLife, 2017, 6: e26248. |
| [14] | Tanner AR, Kennedy VC, Lynch CS, et al. In vivo investigation of ruminant placenta function and physiology—a review [J]. J Anim Sci, 2022, 100(6): skac045. |
| [15] | Qian Q, Xu RY, Wang YP, et al. The NS4A protein of classical swine fever virus suppresses RNA silencing in mammalian cells [J]. J Virol, 2022, 96(15): e01874-21. |
| [16] | Wang DP, Xiu JB, Zhao JY, et al. miR-AB, a miRNA-based shRNA viral toolkit for multicolor-barcoded multiplex RNAi at a single-cell level [J]. EMBO Rep, 2022, 23(4): e53691. |
| [17] | Li Y, Mensah EO, Fordjour E, et al. Recent advances in high-throughput metabolic engineering: Generation of oligonucleotide-mediated genetic libraries [J]. Biotechnol Adv, 2022, 59: 107970. |
| [18] | Boutros M, Ahringer J. The art and design of genetic screens: RNA interference [J]. Nat Rev Genet, 2008, 9(7): 554-566. |
| [19] | Li D, Yang JH, Malik V, et al. An RNAi screen of RNA helicases identifies eIF4A3 as a regulator of embryonic stem cell identity [J]. Nucleic Acids Res, 2022, 50(21): 12462-12479. |
| [20] | Žemaitis K, Subramaniam A, Galeev R, et al. RNAi screen identifies MTA1 as an epigenetic modifier of differentiation commitment in human HSPCs [J]. Exp Hematol, 2022, 115: 20-29. |
| [21] | Wu HY, Lv PY, et al. Genetic screen identified PRMT5 as a neuroprotection target against cerebral ischemia [J]. eLife, 2024, 12: RP89754. |
| [22] | Qi SH, Sun C, Wang J, et al. Identification of NECTIN1 as a novel restriction factor for flavivirus infection [J]. mBio, 2024, 15(12) |
| [23] | Li Q, Wang YH, Hu X, et al. Transcriptional states and chromatin accessibility during bovine myoblasts proliferation and myogenic differentiation [J]. Cell Prolif, 2022, 55(5): e13219. |
| [24] | Wang Q, Zhang QW, Wang XY, et al. Yak FOXO1 and FOXO3 SNPs and association with production traits, and their promotes cells apoptosis via RNAi [J]. Gene, 2020, 743: 144592. |
| [25] | He JN, Liu QY, Yu SY, et al. Expression and functional analysis of the Follistatin-like 3 (FSTL3) gene in the sheep ovary during the oestrous cycle [J]. Reprod Domest Anim, 2021, 56(3): 427-436. |
| [26] | Booker M, Samsonova AA, Kwon Y, et al. False negative rates in Drosophila cell -based-based RNAi screens: a case study [J]. BMC Genom, 2011, 12(1): 50. |
| [27] | Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm [J]. RNA, 2002, 8(7): 855-860. |
| [28] | Grimm D. The dose can make the poison: lessons learned from adverse in vivo toxicities caused by RNAi overexpression [J]. Silence, 2011, 2(1): 8. |
| [29] | Frecot DI, Froehlich T, Rothbauer U. 30 years of nanobodies-an ongoing success story of small binders in biological research [J]. J Cell Sci, 2023, 136(21): jcs261395. |
| [30] | Sone M, Mitsuhashi N, Sugiura Y, et al. Identification of genes supporting cold resistance of mammalian cells: lessons from a hibernator [J]. Cell Death Dis, 2024, 15: 685. |
| [31] | Yamada Y, Miyamoto T, Higuchi S, et al. cDNA expression library screening revealed novel functional genes involved in clear cell carcinogenesis of the ovary in vitro [J]. J Obstet Gynaecol, 2021, 41(1): 100-105. |
| [32] | Zhang P, Kratz AS, Salama M, et al. Expression screening using a Medaka cDNA library identifies evolutionarily conserved regulators of the p53/Mdm2 pathway [J]. BMC Biotechnol, 2015, 15(1): 92. |
| [33] | Mita M. Relaxin-like gonad-stimulating peptides in Asteroidea [J]. Biomolecules, 2023, 13(5): 781. |
| [34] | Kannan S, Miyamoto M, Zhu RJ, et al. Trajectory reconstruction identifies dysregulation of perinatal maturation programs in pluripotent stem cell-derived cardiomyocytes [J]. Cell Rep, 2023, 42(4): 112330. |
| [35] | Ng AHM, Khoshakhlagh P, Rojo Arias JE, et al. A comprehensive library of human transcription factors for cell fate engineering [J]. Nat Biotechnol, 2021, 39(4): 510-519. |
| [36] | Legut M, Gajic Z, Guarino M, et al. A genome-scale screen for synthetic drivers of T cell proliferation [J]. Nature, 2022, 603(7902): 728-735. |
| [37] | 姜光飞, 何维琪, 余蕊, 等. 猪静脉血管内皮细胞cDNA文库构建及与猪非典型瘟病毒云南株关键变异基因互作蛋白的筛选 [J]. 动物医学进展, 2023, 44(7): 1-9. |
| Jiang GF, He WQ, Yu R, et al. Construction of SVEC cDNA library and screening of interacting proteins with key variant gene of APPV-YN [J]. Prog Vet Med, 2023, 44(7): 1-9. | |
| [38] | Wei P, Xue W, Zhao Y, et al. CRISPR-based modular assembly of a UAS-cDNA/ORF plasmid library for more than 5500 Drosophila genes conserved in humans [J]. Genome Res, 2020, 30(1): 95-106. |
| [39] | Tyumentseva M, Tyumentsev A, Akimkin V. CRISPR/Cas9 landscape: current state and future perspectives [J]. Int J Mol Sci, 2023, 24(22): 16077. |
| [40] | Hwang S, Maxwell KL. Diverse mechanisms of CRISPR-Cas9 inhibition by type II anti-CRISPR proteins [J]. J Mol Biol, 2023, 435(7): 168041. |
| [41] | Khoshandam M, Soltaninejad H, Ali Hamidieh A, et al. CRISPR, CAR-T, and NK: Current applications and future perspectives [J]. Genes Dis, 2024, 11(4): 101121. |
| [42] | Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants [J]. Nat Rev Microbiol, 2020, 18(2): 67-83. |
| [43] | Ishibashi R, Maki R, Toyoshima F. Gene targeting in adult organs using in vivo cleavable donor plasmids for CRISPR-Cas9 and CRISPR-Cas12a [J]. Sci Rep, 2024, 14: 7615. |
| [44] | Zhao LX, Qiu MY, Li XJ, et al. CRISPR-Cas13a system: a novel tool for molecular diagnostics [J]. Front Microbiol, 2022, 13: 1060947. |
| [45] | Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells [J]. Nat Rev Genet, 2018, 19(12): 770-788. |
| [46] | Doman JL, Raguram A, Newby GA, et al. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors [J]. Nat Biotechnol, 2020, 38(5): 620-628. |
| [47] | Huang TP, Newby GA, Liu DR. Precision genome editing using cytosine and adenine base editors in mammalian cells [J]. Nat Protoc, 2021, 16(2): 1089-1128. |
| [48] | Agrotis A, Ketteler R. A new age in functional genomics using CRISPR/Cas9 in arrayed library screening [J]. Front Genet, 2015, 6: 300. |
| [49] | Shi YJ, Katdare KA, Kim H, et al. An arrayed CRISPR knockout screen identifies genetic regulators of GLUT1 expression [J]. Sci Rep, 2023, 13: 21038. |
| [50] | Barry T, Mason K, Roeder K, et al. Robust differential expression testing for single-cell CRISPR screens at low multiplicity of infection [J]. Genome Biol, 2024, 25(1): 124. |
| [51] | Shang WJ, Wang F, Fan GF, et al. Key elements for designing and performing a CRISPR/Cas9-based genetic screen [J]. J Genet Genom, 2017, 44(9): 439-449. |
| [52] | Otten ABC, Sun BK. Research techniques made simple: CRISPR genetic screens [J]. J Investig Dermatol, 2020, 140(4): 723-728.e1. |
| [53] | Liang YH, Xie JK, Zhang QJ, et al. AGBE: a dual deaminase-mediated base editor by fusing CGBE with ABE for creating a saturated mutant population with multiple editing patterns [J]. Nucleic Acids Res, 2022, 50(9): 5384-5399. |
| [54] | Zhou YX, Zhu SY, Cai CZ, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells [J]. Nature, 2014, 509(7501): 487-491. |
| [55] | Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening [J]. Nat Meth, 2014, 11(8): 783-784. |
| [56] | Wang XJ, Liu ZW, Li GL, et al. Efficient gene silencing by adenine base editor-mediated start codon mutation [J]. Mol Ther, 2020, 28(2): 431-440. |
| [57] | Liang YJ, Yao XX, Han JX, et al. Establishment of a CRISPR-based lentiviral activation library for transcription factor screening in porcine cells [J]. Animals, 2025, 15(1): 19. |
| [58] | Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9 [J]. Nat Biotechnol, 2016, 34(2): 184-191. |
| [59] | Joung J, Konermann S, Gootenberg JS, et al. Author Correction: Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening [J]. Nat Protoc, 2019, 14(7): 2259. |
| [60] | Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells [J]. Science, 2014, 343(6166): 84-87. |
| [61] | Evers B, Jastrzebski K, Heijmans JPM, et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes [J]. Nat Biotechnol, 2016, 34(6): 631-633. |
| [62] | Morgens DW, Deans RM, Li A, et al. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes [J]. Nat Biotechnol, 2016, 34(6): 634-636. |
| [63] | Merk DJ, Paul L, Tsiami F, et al. CRISPR-Cas9 screens reveal common essential miRNAs in human cancer cell lines [J]. Genome Med, 2024, 16(1): 82. |
| [64] | Su GN, Liu J, Duan CR, et al. Enteric coronavirus PDCoV evokes a non-Warburg effect by hijacking pyruvic acid as a metabolic hub [J]. Redox Biol, 2024, 71: 103112. |
| [65] | Ma NN, Zhang MJ, Zhou JR, et al. Genome-wide CRISPR/Cas9 library screen identifies C16orf62 as a host dependency factor for porcine deltacoronavirus infection [J]. Emerg Microbes Infect, 2024, 13(1): 2400559. |
| [66] | Shen DD, Zhang GG, Weng XG, et al. A genome-wide CRISPR/Cas9 knockout screen identifies TMEM239 as an important host factor in facilitating African swine fever virus entry into early endosomes [J]. PLoS Pathog, 2024, 20(7): e1012256. |
| [67] | 李艳. 基于CRISPR/Cas9文库筛选绒山羊毛囊干细胞增殖必需基因 [D]. 杨凌: 西北农林科技大学, 2024. |
| Li Y. Screening of essential genes for the proliferation of Cashmere goat hair follicle stem cells based on CRISPR/Cas9 library [D]. Yangling: Northwest A & F University, 2024. | |
| [68] | Liu ZX, Dai LL, Sun TH, et al. Massively parallel CRISPR-Cas9 knockout screening in sheep granulosa cells for FSH response genes [J]. Animals, 2024, 14(6): 898. |
| [69] | Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes [J]. Cell, 2013, 154(2): 442-451. |
| [70] | Kampmann M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine [J]. ACS Chem Biol, 2018, 13(2): 406-416. |
| [71] | Xue W, Jian WG, Meng YY, et al. Knockdown of SETD2 promotes erastin-induced ferroptosis in ccRCC [J]. Cell Death Dis, 2023, 14: 539. |
| [72] | Wagner RT, Hlady RA, Pan XY, et al. SETD2 loss-of-function uniquely sensitizes cells to epigenetic targeting of NSD1-directed H3K36 methylation [J]. Genome Biol, 2025, 26(1): 22. |
| [73] | Li E, Benitez C, Boggess SC, et al. CRISPRi-based screens in iAssembloids to elucidate neuron-Glia interactions [J]. Neuron, 2025, 113(5): 701-718.e8. |
| [74] | Kaplan SJ, Wong W, Yan JL, et al. CRISPR screening uncovers a long-range enhancer for ONECUT1 in pancreatic differentiation and links a diabetes risk variant [J]. Cell Rep, 2024, 43(8): 114640. |
| [75] | Zhu XY, Luo H, Yu XR, et al. Genome-wide CRISPRi screening of key genes for recombinant protein expression in Bacillus subtilis [J]. Adv Sci, 2024, 11(33): e2404313. |
| [76] | Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation [J]. Cell, 2014, 159(3): 647-661. |
| [77] | Schmidt R, Steinhart Z, Layeghi M, et al. CRISPR activation and interference screens decode stimulation responses in primary human T cells [J]. Science, 2022, 375(6580): eabj4008. |
| [78] | Kuscu C, Parlak M, Tufan T, et al. CRISPR-STOP gene silencing through base-editing-induced nonsense mutations [J]. Nat Meth, 2017, 14(7): 710-712. |
| [79] | Schmidt R, Ward CC, Dajani R, et al. Base-editing mutagenesis maps alleles to tune human T cell functions [J]. Nature, 2024, 625(7996): 805-812. |
| [80] | Xiao MS, Damodaran AP, Kumari B, et al. Genome-scale exon perturbation screens uncover exons critical for cell fitness [J]. Mol Cell, 2024, 84(13): 2553-2572.e19. |
| [81] | Cheng WS, Liu F, Ren ZJ, et al. Parallel functional assessment of m6A sites in human endodermal differentiation with base editor screens [J]. Nat Commun, 2022, 13: 478. |
| [82] | Li YZ, Xu T, Ma HZ, et al. Functional profiling of serine, threonine and tyrosine sites [J]. Nat Chem Biol, 2025, 21(4): 532-543. |
| [83] | Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases [J]. Nat Biotechnol, 2013, 31(9): 827-832. |
| [84] | Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage [J]. Nature, 2017, 551(7681): 464-471. |
| [85] | Chen SY, Xie WH, Liu ZQ, et al. CRISPR start-loss: a novel and practical alternative for gene silencing through base-editing-induced start codon mutations [J]. Mol Ther Nucleic Acids, 2020, 21: 1062-1073. |
| [86] | Fang L, Wang W, Li GP, et al. CIGAR-seq, a CRISPR/Cas-based method for unbiased screening of novel mRNA modification regulators [J]. Mol Syst Biol, 2020, 16(11): e10025. |
| [87] | Niu Y, Ferreira Azevedo CA, LI X, et al. Multiparametric and accurate functional analysis of genetic sequence variants using CRISPR-Select[J]. Nat Genet, 2022, 54(12): 1983-1993. |
| [88] | Dhainaut M, Rose SA, Akturk G, et al. Spatial CRISPR genomics identifies regulators of the tumor microenvironment [J]. Cell, 2022, 185(7): 1223-1239.e20. |
| [1] | Di Di. Research and Application of Biotechnology in the Field of Veterinary Medicine [J]. Biotechnology Bulletin, 2015, 31(9): 66-69. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||