








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 35-46.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0656
Previous Articles Next Articles
WANG Xin( ), SUN Tao, SUN Mei-li, WANG Kai-feng, JI Xiao-jun(
), SUN Tao, SUN Mei-li, WANG Kai-feng, JI Xiao-jun( )
)
Received:2025-06-23
Online:2025-11-26
Published:2025-12-09
Contact:
JI Xiao-jun
E-mail:xinwang@njtech.edu.cn;xiaojunji@njtech.edu.cn
WANG Xin, SUN Tao, SUN Mei-li, WANG Kai-feng, JI Xiao-jun. Advances in the Biosynthesis of Functional Food Ingredient Hydroxytyrosol[J]. Biotechnology Bulletin, 2025, 41(11): 35-46.
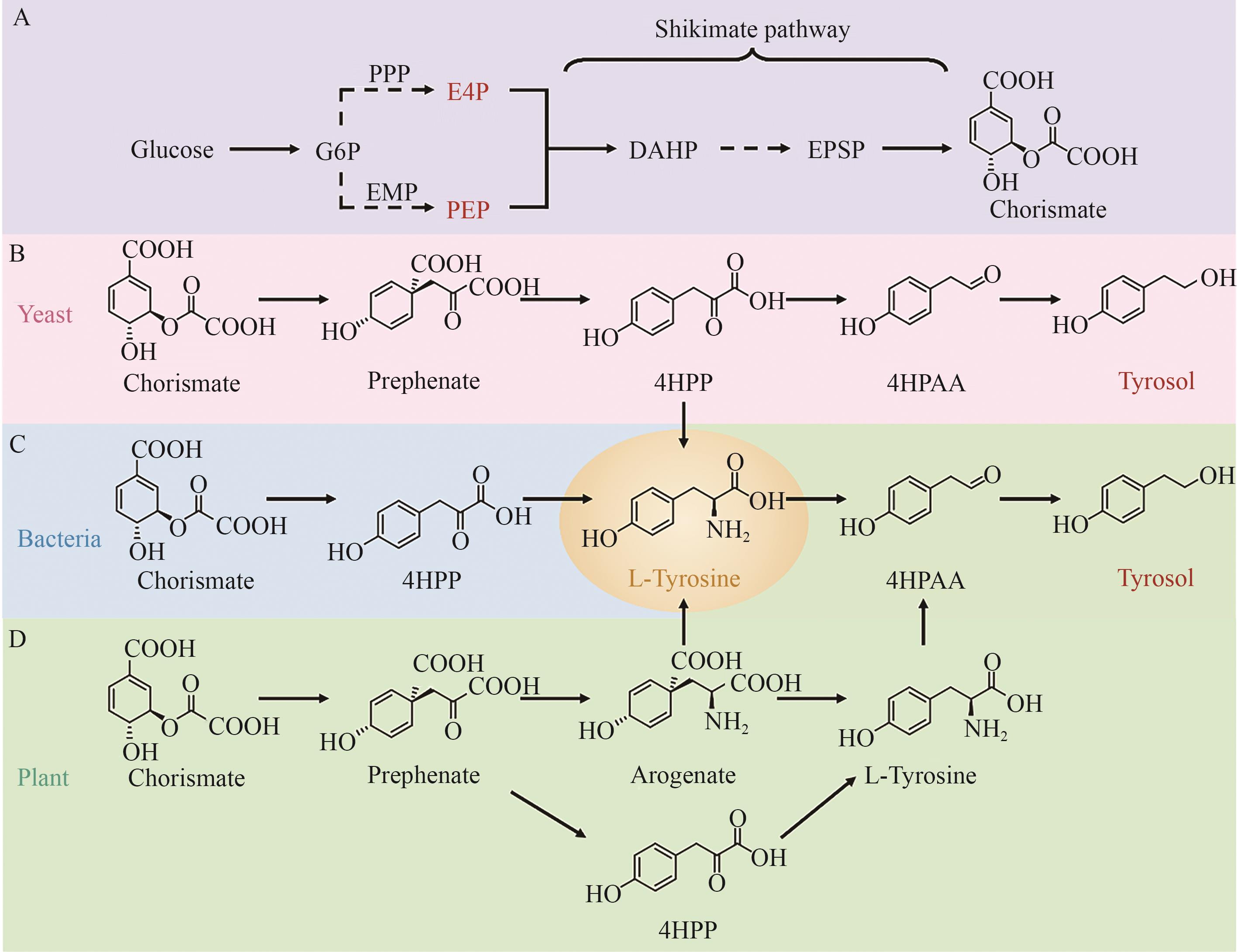
Fig. 2 Biosynthetic pathways of tyrosolA: Biosynthetic pathway from glucose to chorismite. B: Tyrosol biosynthesis from chorismate in yeast. C: L-tyrosine biosynthesis from chorismate in bacteria. D: Tyrosol biosynthesis from chorismate in plants. DAHP: 3-deoxy-D-arabinoheptulosonate-7-phosphate, EPSP: 5-enolpyruvylshikimate-3-phosphatesynthase, E4P: erythrose-4-phosphate, G6P: glucose-6-phosphate, PEP: phosphoenolpyruvate, 4HPAA: 2-(4-hydroxyphenyl) acetaldehyde, 4HPP: 4-hydroxyphenylpyruvic acid
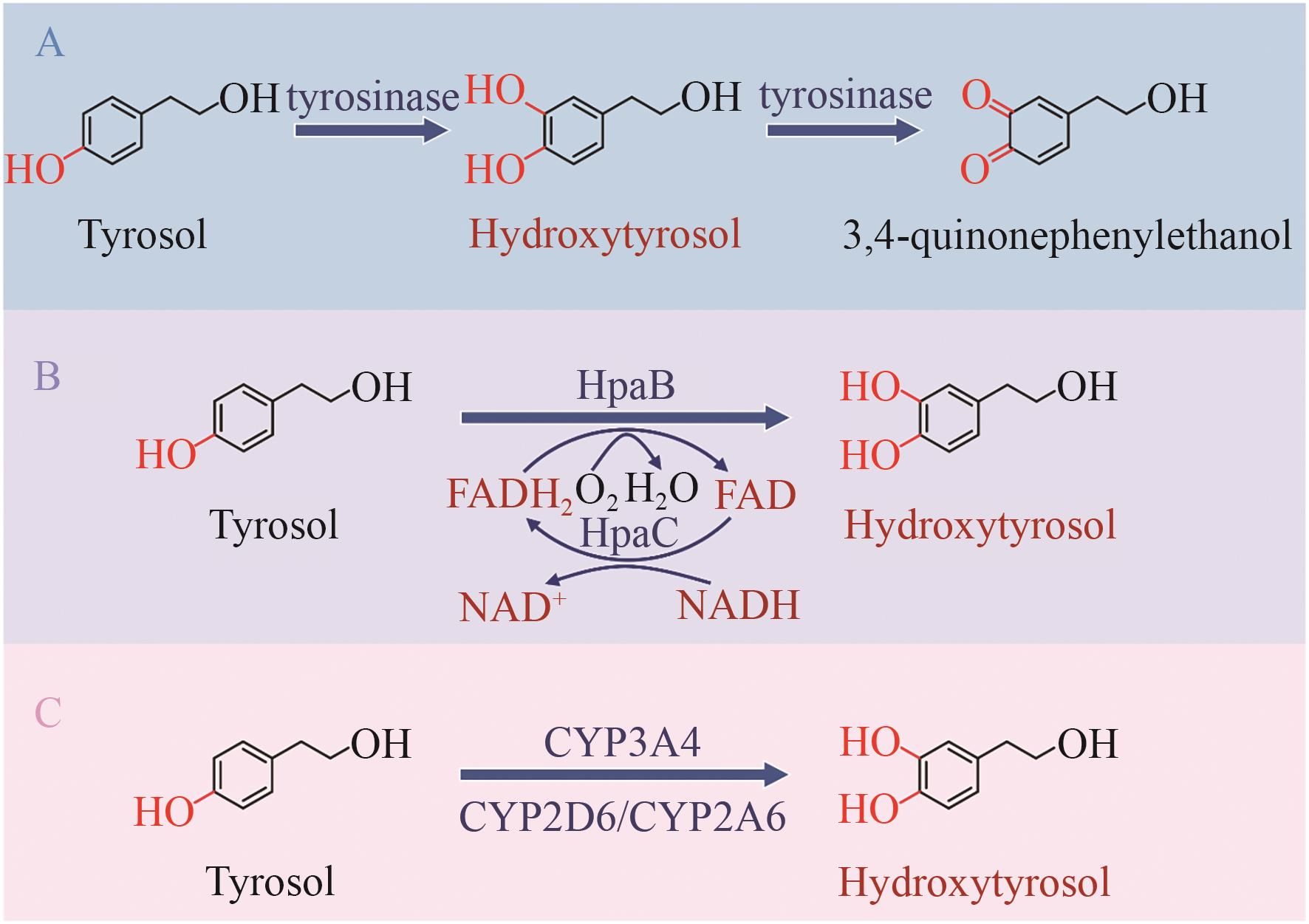
Fig. 3 Biosynthetic pathway from tyrosol to hydroxytyrosolA: Tyrosol is catalyzed by tyrosinase to form hydroxytyrosol, which is further oxidized to 3,4-quinonephenylethanol. B: Tyrosol is converted to hydroxytyrosol via 4-hydroxyphenylacetate 3-hydroxylase. C: Tyrosol is converted to hydroxytyrosol via cytochrome P450 enzymes. CYP3A4/CYP2D6/CYP2A6: Cytochrome P450 enzymes; HpaBC: 4-hydroxyphenylacetate 3-hydroxylase
菌株 Strain | 底物 Substrate | 产量 Titer | 规模 Scale | 合成生物学策略 Metabolic engineering strategies | 参考文献Reference |
|---|---|---|---|---|---|
大肠杆菌 E. coli | 葡萄糖 | 0.08 mmol/L | 摇瓶 | 异源表达鼠来源的TH、人类来源的PCD和DHPR、猪来源的DDC、藤黄微球菌来源的TYO,敲除FEAB | [ |
| 葡萄糖 | 208.00 mg/L | 摇瓶 | 异源表达欧芹来源的AAS,过表达HPABC,敲除TYRR、PHEA和FEAB | [ | |
| 葡萄糖 | 268.30 mg/L | 摇瓶 | 异源表达罂粟来源的TDC和藤黄微球菌来源的TYO,过表达HPABC、AROGfbr 和TYRAfbr,敲除TYRR、 PHEA和FEAB | [ | |
| 甘油,葡萄糖 | (647±35)mg/L | 摇瓶 | 异源表达酿酒酵母来源的ARO10,过表达HPABC和ADH6,敲除FEAB | [ | |
| 葡萄糖 | 270.80 mg/L | 摇瓶 | 异源表达欧芹来源的AAS,青枯雷尔氏菌来源的TYR,过表达ALR-K、 AROA、 AROB、 AROC、 AROD、 AROE、 AROG、 AROL、 PPSA、 TKTA、 TYRA、 TYRB和YDIB | [ | |
| 葡萄糖 | 1.60 g/L | 摇瓶 | 异源表达酿酒酵母来源的ARO10D331C/V,过表达HPABC,敲除FEAB、PHEA、TYRB和TYRR | [ | |
| 葡萄糖 | 8.80 g/L | 3-L发酵罐 | 异源表达巴西固氮螺菌来源的IPDC,过表达AROGfbr 、 PPSA、 TKTA、 AROALC、AROEDB、 TYRAfbr、 YAHK、 HPABC,敲除FEAB、 PYKA、 PHEA、TYRB | [ | |
| 甘油 | 9.87 g/L | 5-L发酵罐 | 异源表达酿酒酵母来源的ARO10和ADH6和奇异变形杆菌来源的LAAD,过表达HPABC、 AROGfbr 、 TYRC,敲除PTSG、 CRR、 PHEA、 TYRR | [ | |
| 葡萄糖 | 4.97 g/L | 5-L发酵罐 | 异源表达解脂耶氏酵母来源的ARO10和PAR4,过表达AROGD146N、 TYRAM53I/A354R 、 HPABC、 GLF、 GLK,敲除PTSG、 PYKA、 MHPB、 PHEA和TYRB | [ | |
酿酒酵母 S. cerevisiae | 蔗糖,甘油 | 308.65 mg/L | 摇瓶 | 异源表达铜绿假单胞菌来源的HPAA/C、欧芹来源的AAS和短双歧杆菌来源的XFPK,过表达ARO4K229L 和ARO7G141S | [ |
| 葡萄糖 | 375.00 mg/L | 摇瓶 | 异源表达大肠杆菌来源的HPABC,过表达ARO4K229L | [ | |
| 葡萄糖 | 6.97 g/L | 5-L发酵罐 | 异源表达铜绿假单胞菌来源的HPAB、大肠杆菌来源的HPAC和TYRAM53I/A354V,过表达ARO3D154N、 ARO4K229L 、 ARO7G141S 、 ARO2、 TKL1、RKL1、 ARO10,敲除PDC1、 PHA2,下调TRP2 | [ | |
| 葡萄糖 | 639.84 mg/L | 摇瓶 | 异源表达大肠杆菌来源的TYRAM53I/A354V 、枯草芽胞杆菌来源的RIBBA、欧芹来源的AAS和短双歧杆菌来源的BbXFPKopt,过表达ARO4K229L 和ARO7G141S,敲除PDC1、 PHA2 | [ | |
酿酒酵母-大肠杆菌 S. cerevisiae- E. coli | 蔗糖 | 435.32 mg/L | 摇瓶 | 酿酒酵母:异源表达欧芹来源的AAS和短双歧杆菌来源的BbXFPKopt,过表达ARO4K229L 、 ARO7G141S;大肠杆菌:过表达HPABC | [ |
地衣芽胞杆菌 B. licheniformis | 葡萄糖 | 9.48 g/L | 5-L发酵罐 | 异源表达大肠杆菌来源的HPABC和乳酸杆菌来源的KIVD,过表达GLCU、 GLCK、 ZWF、 TKT、 AROGfbr 、 AROK、 TYRAfbr、 YUGJ,敲除PYK、 DHBC、 PHEA、 HISG、 DHAS和ADHA | [ |
Table 1 Research progress on de novo synthesis of hydroxytyrosol by microorganisms
菌株 Strain | 底物 Substrate | 产量 Titer | 规模 Scale | 合成生物学策略 Metabolic engineering strategies | 参考文献Reference |
|---|---|---|---|---|---|
大肠杆菌 E. coli | 葡萄糖 | 0.08 mmol/L | 摇瓶 | 异源表达鼠来源的TH、人类来源的PCD和DHPR、猪来源的DDC、藤黄微球菌来源的TYO,敲除FEAB | [ |
| 葡萄糖 | 208.00 mg/L | 摇瓶 | 异源表达欧芹来源的AAS,过表达HPABC,敲除TYRR、PHEA和FEAB | [ | |
| 葡萄糖 | 268.30 mg/L | 摇瓶 | 异源表达罂粟来源的TDC和藤黄微球菌来源的TYO,过表达HPABC、AROGfbr 和TYRAfbr,敲除TYRR、 PHEA和FEAB | [ | |
| 甘油,葡萄糖 | (647±35)mg/L | 摇瓶 | 异源表达酿酒酵母来源的ARO10,过表达HPABC和ADH6,敲除FEAB | [ | |
| 葡萄糖 | 270.80 mg/L | 摇瓶 | 异源表达欧芹来源的AAS,青枯雷尔氏菌来源的TYR,过表达ALR-K、 AROA、 AROB、 AROC、 AROD、 AROE、 AROG、 AROL、 PPSA、 TKTA、 TYRA、 TYRB和YDIB | [ | |
| 葡萄糖 | 1.60 g/L | 摇瓶 | 异源表达酿酒酵母来源的ARO10D331C/V,过表达HPABC,敲除FEAB、PHEA、TYRB和TYRR | [ | |
| 葡萄糖 | 8.80 g/L | 3-L发酵罐 | 异源表达巴西固氮螺菌来源的IPDC,过表达AROGfbr 、 PPSA、 TKTA、 AROALC、AROEDB、 TYRAfbr、 YAHK、 HPABC,敲除FEAB、 PYKA、 PHEA、TYRB | [ | |
| 甘油 | 9.87 g/L | 5-L发酵罐 | 异源表达酿酒酵母来源的ARO10和ADH6和奇异变形杆菌来源的LAAD,过表达HPABC、 AROGfbr 、 TYRC,敲除PTSG、 CRR、 PHEA、 TYRR | [ | |
| 葡萄糖 | 4.97 g/L | 5-L发酵罐 | 异源表达解脂耶氏酵母来源的ARO10和PAR4,过表达AROGD146N、 TYRAM53I/A354R 、 HPABC、 GLF、 GLK,敲除PTSG、 PYKA、 MHPB、 PHEA和TYRB | [ | |
酿酒酵母 S. cerevisiae | 蔗糖,甘油 | 308.65 mg/L | 摇瓶 | 异源表达铜绿假单胞菌来源的HPAA/C、欧芹来源的AAS和短双歧杆菌来源的XFPK,过表达ARO4K229L 和ARO7G141S | [ |
| 葡萄糖 | 375.00 mg/L | 摇瓶 | 异源表达大肠杆菌来源的HPABC,过表达ARO4K229L | [ | |
| 葡萄糖 | 6.97 g/L | 5-L发酵罐 | 异源表达铜绿假单胞菌来源的HPAB、大肠杆菌来源的HPAC和TYRAM53I/A354V,过表达ARO3D154N、 ARO4K229L 、 ARO7G141S 、 ARO2、 TKL1、RKL1、 ARO10,敲除PDC1、 PHA2,下调TRP2 | [ | |
| 葡萄糖 | 639.84 mg/L | 摇瓶 | 异源表达大肠杆菌来源的TYRAM53I/A354V 、枯草芽胞杆菌来源的RIBBA、欧芹来源的AAS和短双歧杆菌来源的BbXFPKopt,过表达ARO4K229L 和ARO7G141S,敲除PDC1、 PHA2 | [ | |
酿酒酵母-大肠杆菌 S. cerevisiae- E. coli | 蔗糖 | 435.32 mg/L | 摇瓶 | 酿酒酵母:异源表达欧芹来源的AAS和短双歧杆菌来源的BbXFPKopt,过表达ARO4K229L 、 ARO7G141S;大肠杆菌:过表达HPABC | [ |
地衣芽胞杆菌 B. licheniformis | 葡萄糖 | 9.48 g/L | 5-L发酵罐 | 异源表达大肠杆菌来源的HPABC和乳酸杆菌来源的KIVD,过表达GLCU、 GLCK、 ZWF、 TKT、 AROGfbr 、 AROK、 TYRAfbr、 YUGJ,敲除PYK、 DHBC、 PHEA、 HISG、 DHAS和ADHA | [ |
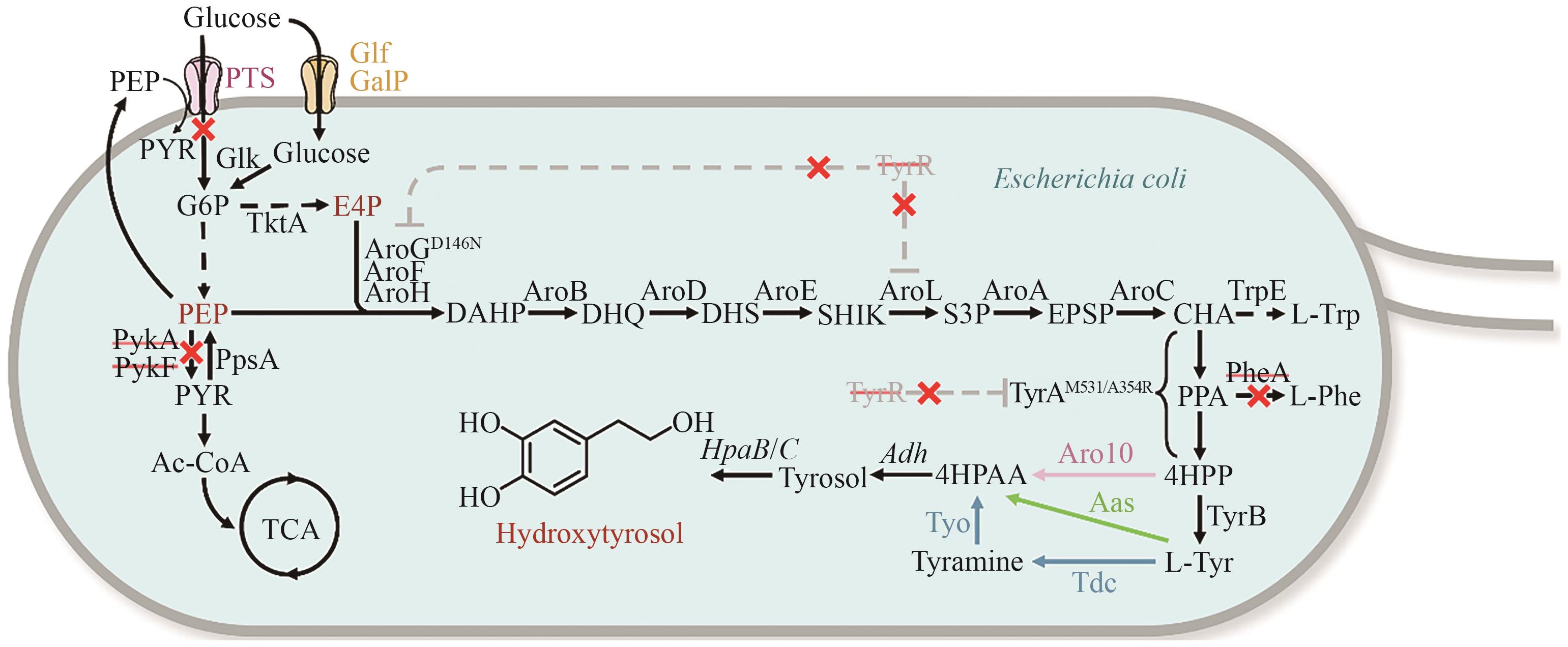
Fig. 4 Synthetic pathway for hydroxytyrosol production in Escherichia coliBlack dashed lines indicate multi-step reactions. Gray dashed lines indicate feedback inhibition. Colored solid lines denote heterologous enzyme-catalyzed steps. Red × indicates gene knockouts. DAHP: 3-deoxy-D-arabinoheptulosonate-7-phosphate, DHQ: 3-dehydroquinate, DHS: 3-dehydroshikimate, EPSP: 5-enolpyruvylshikimate-3-phosphate, E4P: erythrose-4-phosphate, G6P: glucose-6-phosphate, L-Trp: L-tryptophan, L-Tyr: L-Tyrosine, L-Phe: L-phenylalanine, PEP: phosphoenolpyruvate, PPA: prephenate, PYR: pyruvate, SHIK: shikimate,S3P: shikimate-3-phosphate, 4HPAA: 2-(4-hydroxyphenyl) acetaldehyde, 4HPP: 4-hydroxyphenylpyruvic acid
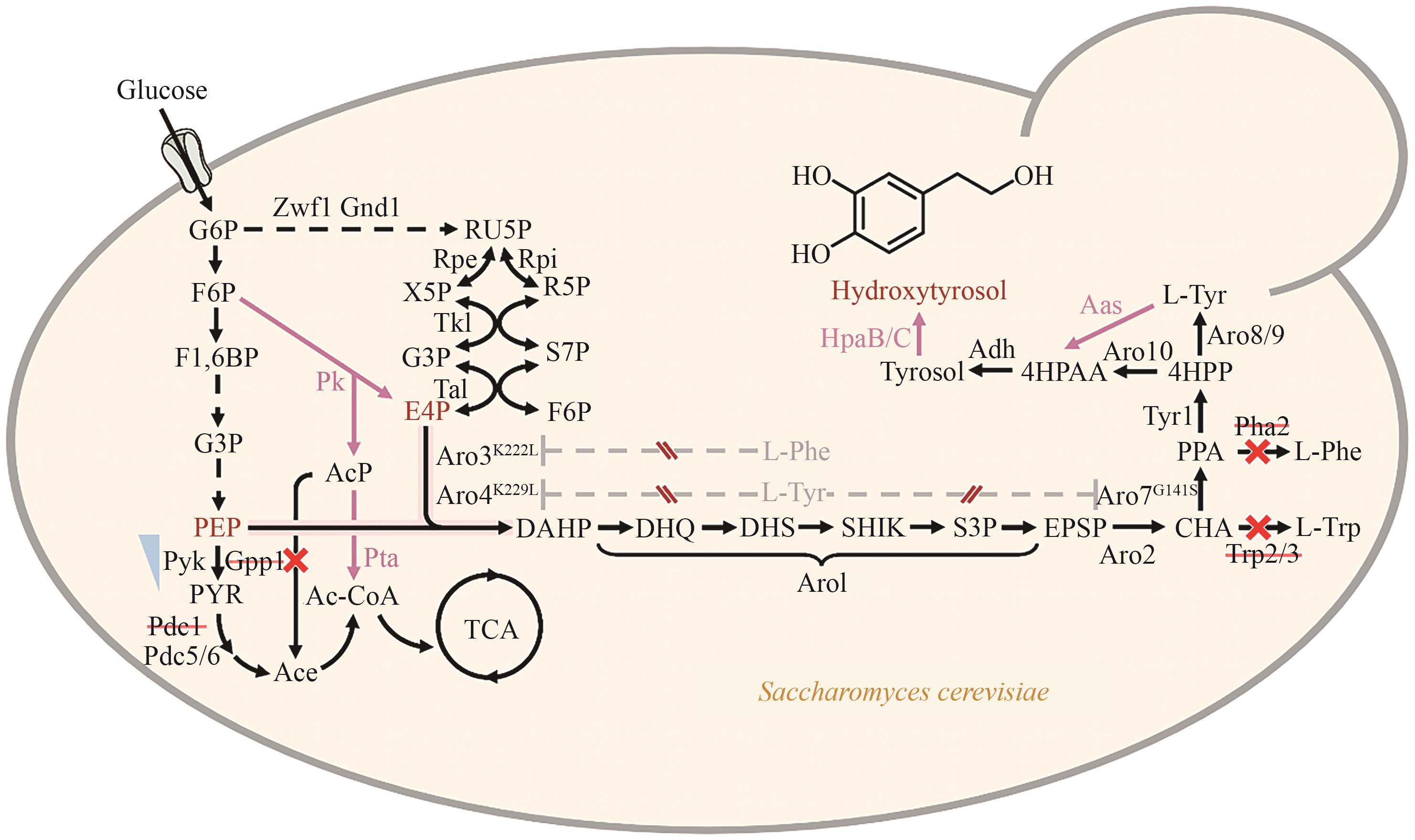
Fig. 5 Synthetic pathway for hydroxytyrosol production in Saccharomyces cerevisiaeBlack dashed lines indicate multi-step reactions. Gray dashed lines indicate feedback inhibition. Red solid lines denote heterologous enzyme-catalyzed steps. Blue inverted triangles indicate downregulation of gene expression. Red × indicates gene knockouts. Ace: acetate, AcP: acetyl phosphate, CHA: chorismate, DAHP: 3-deoxy-D-arabinoheptulosonate-7-phosphate, DHQ: 3-dehydroquinate, DHS: 3-dehydroshikimate, EPSP: 5-enolpyruvylshikimate-3-phosphate, E4P: erythrose-4-phosphate, F1,6BP: fructose-1,6-bisphosphate, F6P: fructose-6-phosphate. G3P: glyceraldehyde-3-phosphate, G6P: glucose-6-phosphate, L-Trp: L-Tryptophan, L-Tyr: L-Tyrosine, L-Phe: L-Phenylalanine, PEP: phosphoenolpyruvate, PPA: prephenate, PYR: pyruvate, RU5P: ribulose-5-phosphate, R5P: ribose-5-phosphate, SHIK: shikimate, S3P: shikimate-3-phosphate, S7P: sedoheptulose-7-phosphate, X5P: xylulose-5-phosphate, 4HPAA: 2-(4-hydroxyphenyl) acetaldehyde, 4HPP: 4-hydroxyphenylpyruvic acid
| [1] | Gallardo-Fernández M, Gonzalez-Ramirez M, Cerezo AB, et al. Hydroxytyrosol in foods: analysis, food sources, EU dietary intake, and potential uses [J]. Foods, 2022, 11(15): 2355. |
| [2] | Gonzalez-Ramirez M, Gallardo-Fernandez M, Cerezo AB, et al. The production of bioactive hydroxytyrosol in fermented beverages: the role of must composition and a genetically modified yeast strain [J]. Fermentation, 2024, 10(4): 198. |
| [3] | Mastralexi A, Nenadis N, Tsimidou MZ. Addressing analytical requirements to support health claims on “olive oil polyphenols” (EC Regulation 432/2012) [J]. J Agric Food Chem, 2014, 62(12): 2459-2461. |
| [4] | Visioli F, Poli A, Gall C. Antioxidant and other biological activities of phenols from olives and olive oil [J]. Med Res Rev, 2002, 22(1): 65-75. |
| [5] | Servili M, Rizzello CG, Taticchi A, et al. Functional milk beverage fortified with phenolic compounds extracted from olive vegetation water, and fermented with functional lactic acid bacteria [J]. Int J Food Microbiol, 2011, 147(1): 45-52. |
| [6] | Bañares C, Martin D, Reglero G, et al. Protective effect of hydroxytyrosol and rosemary extract in a comparative study of the oxidative stability of Echium oil [J]. Food Chem, 2019, 290: 316-323. |
| [7] | Raposo R, Ruiz-Moreno MJ, Garde-Cerdán T, et al. Effect of hydroxytyrosol on quality of sulfur dioxide-free red wine [J]. Food Chem, 2016, 192: 25-33. |
| [8] | Medina-Martínez MS, Truchado P, Castro-Ibáñez I, et al. Antimicrobial activity of hydroxytyrosol: a current controversy [J]. Biosci Biotechnol Biochem, 2016, 80(4): 801-810. |
| [9] | Bedoya LM, Beltrán M, Obregón-Calderón P, et al. Hydroxytyrosol: a new class of microbicide displaying broad anti-HIV-1 activity [J]. AIDS, 2016, 30(18): 2767-2776. |
| [10] | Chaves-López C, Serio A, Mazzarrino G, et al. Control of household mycoflora in fermented sausages using phenolic fractions from olive mill wastewaters [J]. Int J Food Microbiol, 2015, 207: 49-56. |
| [11] | Bertelli M, Kiani AK, Paolacci S, et al. Hydroxytyrosol: a natural compound with promising pharmacological activities [J]. J Biotechnol, 2020, 309: 29-33. |
| [12] | Kalogerakis N, Politi M, Foteinis S, et al. Recovery of antioxidants from olive mill wastewaters: a viable solution that promotes their overall sustainable management [J]. J Environ Manag, 2013, 128: 749-758. |
| [13] | Ziosi P, Paolucci C, Santarelli F, et al. A two-step process for the synthesis of hydroxytyrosol [J]. ChemSusChem, 2018, 11(13): 2202-2210. |
| [14] | Bovicelli P, Antonioletti R, Mancini S, et al. Expedient synthesis of hydroxytyrosol and its esters [J]. Synth Commun, 2007, 37(23): 4245-4252. |
| [15] | Tang JL, Wang JY, Gong PF, et al. Biosynthesis and biotechnological synthesis of hydroxytyrosol [J]. Foods, 2024, 13(11): 1694. |
| [16] | Jiang M, Zhang HR. Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli [J]. Curr Opin Biotechnol, 2016, 42: 1-6. |
| [17] | Schenck CA, Maeda HA. Tyrosine biosynthesis, metabolism, and catabolism in plants [J]. Phytochemistry, 2018, 149: 82-102. |
| [18] | Cordente AG, Schmidt S, Beltran G, et al. Harnessing yeast metabolism of aromatic amino acids for fermented beverage bioflavouring and bioproduction [J]. Appl Microbiol Biotechnol, 2019, 103(11): 4325-4336. |
| [19] | Hazelwood LA, Daran JM, van Maris AJA, et al. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism [J]. Appl Environ Microbiol, 2008, 74(8): 2259-2266. |
| [20] | 孙中贯, 刘琳, 王亚平, 等. 酿酒酵母高级醇代谢研究进展 [J]. 生物工程学报, 2021, 37(2): 429-447. |
| Sun ZG, Liu L, Wang YP, et al. Higher alcohols metabolism by Saccharomyces cerevisiae: a mini review [J]. Chin J Biotechnol, 2021, 37(2): 429-447. | |
| [21] | Nisbet MA, Tobias HJ, Brenna JT, et al. Quantifying the contribution of grape hexoses to wine volatiles by high-precision [U¹³C]-glucose tracer studies [J]. J Agric Food Chem, 2014, 62(28): 6820-6827. |
| [22] | Claus H, Decker H. Bacterial tyrosinases [J]. Syst Appl Microbiol, 2006, 29(1): 3-14. |
| [23] | Olivares C, Solano F. New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins [J]. Pigment Cell Melanoma Res, 2009, 22(6): 750-760. |
| [24] | Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases [J]. Chem Rev, 1996, 96(7): 2563-2606. |
| [25] | Deri-Zenaty B, Bachar S, Rebroš M, et al. A coupled enzymatic reaction of tyrosinase and glucose dehydrogenase for the production of hydroxytyrosol [J]. Appl Microbiol Biotechnol, 2020, 104(11): 4945-4955. |
| [26] | Rodríguez-Morató J, Robledo P, Tanner JA, et al. CYP2D6 and CYP2A6 biotransform dietary tyrosol into hydroxytyrosol [J]. Food Chem, 2017, 217: 716-725. |
| [27] | Satoh Y, Tajima K, Munekata M, et al. Engineering of l-tyrosine oxidation in Escherichia coli and microbial production of hydroxytyrosol [J]. Metab Eng, 2012, 14(6): 603-610. |
| [28] | Chung D, Kim SY, Ahn JH. Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli [J]. Sci Rep, 2017, 7(1): 2578. |
| [29] | Choo HJ, Kim EJ, Kim SY, et al. Microbial synthesis of hydroxytyrosol and hydroxysalidroside [J]. Appl Biol Chem, 2018, 61(3): 295-301. |
| [30] | Li XL, Chen ZY, Wu YF, et al. Establishing an artificial pathway for efficient biosynthesis of hydroxytyrosol [J]. ACS Synth Biol, 2018, 7(2): 647-654. |
| [31] | Trantas E, Navakoudis E, Pavlidis T, et al. Dual pathway for metabolic engineering of Escherichia coli to produce the highly valuable hydroxytyrosol [J]. PLoS One, 2019, 14(11): e0212243. |
| [32] | Xia YY, Qi LN, Shi XL, et al. Construction of an Escherichia coli cell factory for de novo synthesis of tyrosol through semi-rational design based on phenylpyruvate decarboxylase ARO10 engineering [J]. Int J Biol Macromol, 2023, 253: 127385. |
| [33] | Koma D, Fujisawa M, Ohashi H, et al. Production of 3-hydroxytyrosol from glucose by chromosomally engineered Escherichia coli by fed-batch cultivation in a jar fermenter [J]. J Agric Food Chem, 2023, 71(24): 9451-9459. |
| [34] | Wang HJ, Wang L, Chen JB, et al. Promoting FADH2 regeneration of hydroxylation for high-level production of hydroxytyrosol from glycerol in Escherichia coli [J]. J Agric Food Chem, 2023, 71(44): 16681-16690. |
| [35] | Chen XC, Qian T, Wei WP, et al. De novo synthesis of tyrosol and hydroxytyrosol through temperature-inducible systems and metabolic engineering [J]. ACS Synth Biol, 2025, 14(6): 2294-2304. |
| [36] | Liu YJ, Liu H, Hu HT, et al. De novo production of hydroxytyrosol by metabolic engineering of Saccharomyces cerevisiae [J]. J Agric Food Chem, 2022, 70(24): 7490-7499. |
| [37] | Bisquert R, Planells-Cárcel A, Valera-García E, et al. Metabolic engineering of Saccharomyces cerevisiae for hydroxytyrosol overproduction directly from glucose [J]. Microb Biotechnol, 2022, 15(5): 1499-1510. |
| [38] | Liu HY, Wu XX, Ma H, et al. High-level production of hydroxytyrosol in engineered Saccharomyces cerevisiae [J]. ACS Synth Biol, 2022, 11(11): 3706-3713. |
| [39] | Liu YJ, Gu BX, Shi JH, et al. Inverse metabolic engineering based on metabonomics for efficient production of hydroxytyrosol by Saccharomyces cerevisiae [J]. Bioresour Technol, 2024, 409: 131187. |
| [40] | Liu YJ, Song D, Hu HT, et al. De novo production of hydroxytyrosol by Saccharomyces cerevisiae-Escherichia coli coculture engineering [J]. ACS Synth Biol, 2022, 11(9): 3067-3077. |
| [41] | Zhan YY, Zhou F, Ruan WQ, et al. Systematic metabolic engineering of Bacillus licheniformis for hyperproduction of the antioxidant hydroxytyrosol [J]. Green Chem, 2023, 25(21): 8718-8729. |
| [42] | Yang D, Park SY, Park YS, et al. Metabolic engineering of Escherichia coli for natural product biosynthesis [J]. Trends Biotechnol, 2020, 38(7): 745-765. |
| [43] | Iacometti C, Marx K, Hönick M, et al. Activating silent glycolysis bypasses in Escherichia coli [J]. Biodes Res, 2022, 2022: 9859643. |
| [44] | Liu XZ, Niu H, Li Q, et al. Metabolic engineering for the production of l-phenylalanine in Escherichia coli [J]. 3 Biotech, 2019, 9(3): 85. |
| [45] | Siedler S, Bringer S, Blank LM, et al. Engineering yield and rate of reductive biotransformation in Escherichia coli by partial cyclization of the pentose phosphate pathway and PTS-independent glucose transport [J]. Appl Microbiol Biotechnol, 2012, 93(4): 1459-1467. |
| [46] | Liu YF, Xu YR, Ding DQ, et al. Genetic engineering of Escherichia coli to improve L-phenylalanine production [J]. BMC Biotechnol, 2018, 18(1): 5. |
| [47] | Chandran SS, Yi J, Draths KM, et al. Phosphoenolpyruvate availability and the biosynthesis of shikimic acid [J]. Biotechnol Prog, 2003, 19(3): 808-814. |
| [48] | 盛华康, 张博, 申晓林, 等. 微生物合成白藜芦醇及其衍生物[J]. 化工进展, 2025, 44(5): 2463-2474. |
| Sheng HK, Zhang B, Shen XL, et al. Microbial synthesis of resveratrol and its derivatives [J]. Chem Ind Eng Prog, 2025, 44(5): 2463-2474. | |
| [49] | Koma D, Kishida T, Yoshida E, et al. Chromosome engineering to generate plasmid-free phenylalanine- and tyrosine-overproducing Escherichia coli strains that can be applied in the generation of aromatic-compound-producing bacteria [J]. Appl Environ Microbiol, 2020, 86(14): e00525-20. |
| [50] | 江晶洁, 刘涛, 林双君. 基于莽草酸途径微生物合成芳香族化合物及其衍生物的研究进展 [J]. 生命科学, 2019, 31(5): 430-448. |
| Jiang JJ, Liu T, Lin SJ. Research progress on the biosynthesis of aromatic compounds by microorganisms [J]. Chin Bull Life Sci, 2019, 31(5): 430-448. | |
| [51] | Cui D, Deng AH, Bai H, et al. Molecular basis for feedback inhibition of tyrosine-regulated 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli [J]. J Struct Biol, 2019, 206(3): 322-334. |
| [52] | Rodriguez A, Martínez JA, Flores N, et al. Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds [J]. Microb Cell Fact, 2014, 13(1): 126. |
| [53] | Pittard J, Camakaris H, Yang J. The TyrR regulon [J]. Mol Microbiol, 2005, 55(1): 16-26. |
| [54] | Bai YF, Bi HP, Zhuang YB, et al. Production of salidroside in metabolically engineered Escherichia coli [J]. Sci Rep, 2014, 4: 6640. |
| [55] | Satoh Y, Tajima K, Munekata M, et al. Engineering of a tyrosol-producing pathway, utilizing simple sugar and the central metabolic tyrosine, in Escherichia coli [J]. J Agric Food Chem, 2012, 60(4): 979-984. |
| [56] | Chen W, Yao J, Meng J, et al. Promiscuous enzymatic activity-aided multiple-pathway network design for metabolic flux rearrangement in hydroxytyrosol biosynthesis [J]. Nat Commun, 2019, 10(1): 960. |
| [57] | Kaminaga Y, Schnepp J, Peel G, et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation [J]. J Biol Chem, 2006, 281(33): 23357-23366. |
| [58] | Mermigka G, Vavouraki AI, Nikolaou C, et al. An engineered plant metabolic pathway results in high yields of hydroxytyrosol due to a modified whole-cell biocatalysis in bioreactor [J]. Metabolites, 2023, 13(11): 1126. |
| [59] | Zhao Y, Coelho C, Lauer S, et al. CREEPY: CRISPR-mediated editing of synthetic episomes in yeast [J]. Nucleic Acids Res, 2023, 51(13): e72. |
| [60] | Lian JZ, Mishra S, Zhao HM. Recent advances in metabolic engineering of Saccharomyces cerevisiae: New tools and their applications [J]. Metab Eng, 2018, 50: 85-108. |
| [61] | Borodina I, Nielsen J. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals [J]. Biotechnol J, 2014, 9(5): 609-620. |
| [62] | Suástegui M, Guo WH, Feng XY, et al. Investigating strain dependency in the production of aromatic compounds in Saccharomyces cerevisiae [J]. Biotechnol Bioeng, 2016, 113(12): 2676-2685. |
| [63] | Paravicini G, Schmidheini T, Braus G. Purification and properties of the 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (phenylalanine-inhibitable) of Saccharomyces cerevisiae [J]. Eur J Biochem, 1989, 186(1-2): 361-366. |
| [64] | Schnappauf G, Hartmann M, Künzler M, et al. The two 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase isoenzymes from Saccharomyces cerevisiae show different kinetic modes of inhibition [J]. Arch Microbiol, 1998, 169(6): 517-524. |
| [65] | Yang SL, Pan XW, You JJ, et al. Systematic metabolic engineering of Yarrowia lipolytica for the enhanced production of erythritol [J]. Bioresour Technol, 2024, 391: 129918. |
| [66] | Suástegui M, Yu Ng C, Chowdhury A, et al. Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in Saccharomyces cerevisiae for high production of polymer and drug precursors [J]. Metab Eng, 2017, 42: 134-144. |
| [67] | Deaner M, Alper HS. Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae [J]. Metab Eng, 2017, 40: 14-22. |
| [68] | Liu QL, Yu T, Li XW, et al. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals [J]. Nat Commun, 2019, 10(1): 4976. |
| [69] | Castaño-Cerezo S, Pastor JM, Renilla S, et al. An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli [J]. Microb Cell Fact, 2009, 8: 54. |
| [70] | Guo W, Huang QL, Feng YH, et al. Rewiring central carbon metabolism for tyrosol and salidroside production in Saccharomyces cerevisiae [J]. Biotechnol Bioeng, 2020, 117(8): 2410-2419. |
| [71] | Song N, Xia HL, Xie YR, et al. Semi-rational design and modification of phosphoketolase to improve the yield of tyrosol in Saccharomyces cerevisiae [J]. Synth Syst Biotechnol, 2025, 10(1): 294-306. |
| [72] | Gold ND, Gowen CM, Lussier FX, et al. Metabolic engineering of a tyrosine-overproducing yeast platform using targeted metabolomics [J]. Microb Cell Fact, 2015, 14: 73. |
| [73] | Brochado AR, Matos C, Møller BL, et al. Improved vanillin production in baker’s yeast through in silico design [J]. Microb Cell Fact, 2010, 9: 84. |
| [74] | Remize F, Andrieu E, Dequin S. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation [J]. Appl Environ Microbiol, 2000, 66(8): 3151-3159. |
| [75] | Guo W, Huang QL, Liu H, et al. Rational engineering of chorismate-related pathways in Saccharomyces cerevisiae for improving tyrosol production [J]. Front Bioeng Biotechnol, 2019, 7: 152. |
| [76] | Sträter N, Schnappauf G, Braus G, et al. Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures [J]. Structure, 1997, 5(11): 1437-1452. |
| [77] | Brückner C, Oreb M, Kunze G, et al. An expanded enzyme toolbox for production of cis, cis-muconic acid and other shikimate pathway derivatives in Saccharomyces cerevisiae [J]. FEMS Yeast Res, 2018, 18(2). DOI: 10.1093/femsyr/foy017 . |
| [78] | Strater N, Hakansson K, Schnappauf G, et al. Crystal structure of the T state of allosteric yeast chorismate mutase and comparison with the R state [J]. Proc Natl Acad Sci USA, 1996, 93(8): 3330-3334. |
| [79] | Luttik MAH, Vuralhan Z, Suir E, et al. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: Quantification of metabolic impact [J]. Metab Eng, 2008, 10(3/4): 141-153. |
| [80] | Liu HY, Xiao QJ, Wu XX, et al. Mechanistic investigation of a D to N mutation in DAHP synthase that dictates carbon flux into the shikimate pathway in yeast [J]. Commun Chem, 2023, 6(1): 152. |
| [81] | Kallscheuer N, Menezes R, Foito A, et al. Identification and microbial production of the raspberry phenol salidroside that is active against Huntington’s disease [J]. Plant Physiol, 2019, 179(3): 969-985. |
| [82] | Rodriguez A, Kildegaard KR, Li MJ, et al. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis [J]. Metab Eng, 2015, 31: 181-188. |
| [83] | Anissi J, Sendide K, Ouardaoui A, et al. Production of hydroxytyrosol from hydroxylation of tyrosol by Rhodococcus pyridinivorans 3HYL DSM109178 [J]. Biocatal Biotransform, 2021, 39(6): 418-428. |
| [84] | Gu Y, Ma JB, Zhu YL, et al. Engineering Yarrowia lipolytica as a chassis for De novo synthesis of five aromatic-derived natural products and chemicals [J]. ACS Synth Biol, 2020, 9(8): 2096-2106. |
| [85] | Gu Y, Ma JB, Zhu YL, et al. Refactoring Ehrlich pathway for high-yield 2-phenylethanol production in Yarrowia lipolytica [J]. ACS Synth Biol, 2020, 9(3): 623-633. |
| [86] | Guan AL, He ZX, Wang X, et al. Engineering the next-generation synthetic cell factory driven by protein engineering [J]. Biotechnol Adv, 2024, 73: 108366. |
| [87] | Chen ZY, Wu T, Yu SZ, et al. Self-assembly systems to troubleshoot metabolic engineering challenges [J]. Trends Biotechnol, 2024, 42(1): 43-60. |
| [1] | HE Yu-bing, FU Zhen-hao, LI Ren-han, LIU Xiu-xia, LIU Chun-li, YANG Yan-kun, LI Ye, BAI Zhong-hu. Efficient Biosynthesis of 2-Naphthaleneethanol in Metabolically Engineered Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2024, 40(7): 99-107. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||