








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 14-21.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0691
Received:2025-06-30
Online:2025-11-26
Published:2025-12-09
Contact:
ZHOU Yong-jin
E-mail:baifan1215@dicp.ac.cn;zhouyongjin@dicp.ac.cn
BAI Fan, ZHOU Yong-jin. Yeast Single-cell Proteins Promote the Development and Application of Food Industry[J]. Biotechnology Bulletin, 2025, 41(11): 14-21.
菌种 Species | 发酵底物 Fermentation substrate | 蛋白质含量 Protein content (%) | 参考文献 Reference |
|---|---|---|---|
| S. cerevisiae | 橙浆和废谷物 | 38.5 | [ |
| D. hansenii | 啤酒厂废谷物 | 31.8 | [ |
| K. marxianus | 橙浆、废谷物、马铃薯片等 | 33.7 | [ |
| 干酪乳清 | 43.4 | [ | |
| Y. lipolytica | 食品工业废物 | 38.8 | [ |
| C. utilis | 马铃薯淀粉工业废物 | 46.1 | [ |
| K. phaffii | 甲醇 | 50.6 | [ |
| 甲醇 | 67.2 | [ |
Table 1 Production of single-cell proteins using various food and industrial wastes by yeast cells
菌种 Species | 发酵底物 Fermentation substrate | 蛋白质含量 Protein content (%) | 参考文献 Reference |
|---|---|---|---|
| S. cerevisiae | 橙浆和废谷物 | 38.5 | [ |
| D. hansenii | 啤酒厂废谷物 | 31.8 | [ |
| K. marxianus | 橙浆、废谷物、马铃薯片等 | 33.7 | [ |
| 干酪乳清 | 43.4 | [ | |
| Y. lipolytica | 食品工业废物 | 38.8 | [ |
| C. utilis | 马铃薯淀粉工业废物 | 46.1 | [ |
| K. phaffii | 甲醇 | 50.6 | [ |
| 甲醇 | 67.2 | [ |
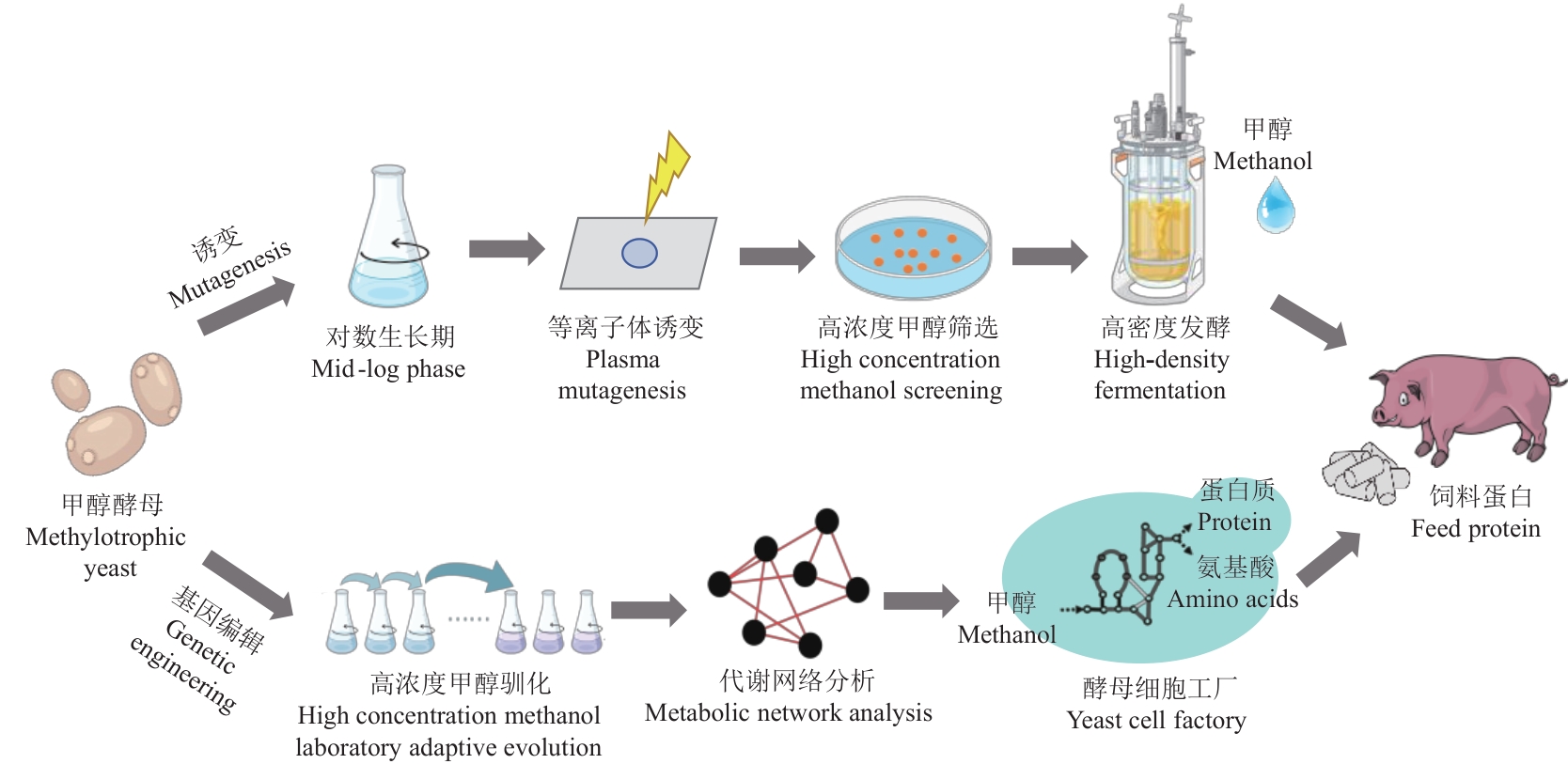
Fig. 1 Strategy of producing single-cell proteins by methylotrophic yeastThere are two strategies of bioconversion methanol to produce single-cell proteins by unconventional yeast species, one is traditional mutagenesis method, methylotrophic yeast was mutagenized and screened on high methanol concentration plate, and the mutated strains utilizing high concentration methanol are selected and performed industrial fermentation to produce single-cell proteins as feed protein additives. Another strategy is genetic editing, methylotrophic yeasts are proceeded through high-concentration methanol of laboratory adaptive evolution, the discrepancy of metabolic network are systematically analyzed in methanol-evolved strain, the potential methanol utilization genes target are explored, and modification of metabolic engineering is conducted, and finally the engineering strains efficiently utilizing methanol to produce proteins are used for producing single-cell proteins
菌株 Species | 改造方式 Modifying methods | 改造应用 Modifing application | 参考文献 References |
|---|---|---|---|
| S. pastorianus | UV诱变 | 减少乙醛生产 | [ |
| UV/EMS诱变 | 提高对高重力麦芽汁发酵 | [ | |
| EMS诱变 | 提高对热击耐受 | [ | |
| 干扰MET10、SKP2表达,过表达HOM3 | 提高SO2生产,减少H2S生产 | [ | |
| 过表达AGT1 | 提高麦芽糖利用 | [ | |
| S. cerevisiae | EMS诱变 | 提高对乙醇耐受 | [ |
| UV诱变 | 减少H2S生产,提高谷胱甘肽生产 | [ | |
| 过表达FLO1、FLO5、FLO11 | 增强絮凝 | [ | |
| 干扰PEP4表达 | 提高泡沫稳定性 | [ | |
| 表达异源基因 | 增加香草醛风味 | [ | |
| 表达异源基因 | 增加树莓酮风味 | [ | |
| K. phaffii | 表达异源基因 | 生产血红素类蛋白 | [ |
| 增加LegH拷贝数 | 生产大豆血红蛋白 | [ |
Table 2 Enriching the functions of yeast single-cell proteins by different modifing methods
菌株 Species | 改造方式 Modifying methods | 改造应用 Modifing application | 参考文献 References |
|---|---|---|---|
| S. pastorianus | UV诱变 | 减少乙醛生产 | [ |
| UV/EMS诱变 | 提高对高重力麦芽汁发酵 | [ | |
| EMS诱变 | 提高对热击耐受 | [ | |
| 干扰MET10、SKP2表达,过表达HOM3 | 提高SO2生产,减少H2S生产 | [ | |
| 过表达AGT1 | 提高麦芽糖利用 | [ | |
| S. cerevisiae | EMS诱变 | 提高对乙醇耐受 | [ |
| UV诱变 | 减少H2S生产,提高谷胱甘肽生产 | [ | |
| 过表达FLO1、FLO5、FLO11 | 增强絮凝 | [ | |
| 干扰PEP4表达 | 提高泡沫稳定性 | [ | |
| 表达异源基因 | 增加香草醛风味 | [ | |
| 表达异源基因 | 增加树莓酮风味 | [ | |
| K. phaffii | 表达异源基因 | 生产血红素类蛋白 | [ |
| 增加LegH拷贝数 | 生产大豆血红蛋白 | [ |
| [1] | Pereira AG, Fraga-Corral M, Garcia-Oliveira P, et al. Single-cell proteins obtained by circular economy intended as a feed ingredient in aquaculture [J]. Foods, 2022, 11(18): 2831. |
| [2] | Lapeña D, Kosa G, Hansen LD, et al. Production and characterization of yeasts grown on media composed of spruce-derived sugars and protein hydrolysates from chicken by-products [J]. Microb Cell Fact, 2020, 19(1): 19. |
| [3] | Meng J, Liu SF, Gao L, et al. Economical production of Pichia pastoris single cell protein from methanol at industrial pilot scale [J]. Microb Cell Fact, 2023, 22(1): 198. |
| [4] | Liu KY, Huang SY, Zhang L, et al. Efficient production of single cell protein from biogas slurry using screened alkali-salt-tolerant Debaryomyces hansenii [J]. Bioresour Technol, 2024, 393: 130119. |
| [5] | Bratosin BC, Darjan S, Vodnar DC. Single cell protein: a potential substitute in human and animal nutrition [J]. Sustainability, 2021, 13(16): 9284. |
| [6] | Zhao Y, Han ZW, Zhu XC, et al. Yeast proteins: proteomics, extraction, modification, functional characterization, and structure: a review [J]. J Agric Food Chem, 2024, 72(34): 18774-18793. |
| [7] | 黄庆生. 我国粮食安全战略下饲料粮保供策略思考 [J]. 中国饲料, 2023, 1(22): 15-25. |
| Huang QS. Thoughts on feed grain supply assurance strategies under China’s food security strategy [J]. China Feed, 2023, 1(22): 15-25. | |
| [8] | Lambo MT, Ma HK, Zhang HS, et al. Mechanism of action, benefits, and research gap in fermented soybean meal utilization as a high-quality protein source for livestock and poultry [J]. Anim Nutr, 2024, 16: 130-146. |
| [9] | 吴信, 张会会, 杨旭东, 等. 新型单细胞蛋白饲料资源开发利用进展 [J]. 饲料工业, 2024, 45 (20): 1-8. |
| Wu X, Zhang HH, Yang XD, et al. Progress in the development and utilization of new feed resources of single cell protein [J]. Feed Industry, 2024, 45 (20): 1-8. | |
| [10] | 张博, 杨晓伟, 康志勇,等. 饲用豆粕减量替代背景下杂粕型饲料蛋白资源开发应用研究进展 [J]. 中国饲料, 2025, (9): 146-160. |
| Zhang B, Yang XW, Kang ZY, et al. Research progress on development and application of miscellaneous meal feed protein resources under the background of soybean meal reduction substitution [J]. China Feed, 2025, (9): 146-160. | |
| [11] | Zhang QL, Liang HL, Longshaw M, et al. Effects of replacing fishmeal with methanotroph (Methylococcus capsulatus, Bath) bacteria meal (FeedKind®) on growth and intestinal health status of juvenile largemouth bass (Micropterus salmoides) [J]. Fish Shellfish Immunol, 2022, 122: 298-305. |
| [12] | Zhuang ZK, Wan GY, Lu XC, et al. Metabolic engineering for single-cell protein production from renewable feedstocks and its applications [J]. Adv Biotechnol, 2024, 2(4): 35. |
| [13] | Jach ME, Serefko A, Ziaja M, et al. Yeast protein as an easily accessible food source [J]. Metabolites, 2022, 12(1): 63. |
| [14] | Anderson A, Van der Mijnsbrugge A, Cameleyre X, et al. From yeast screening for suitability as single cell protein to fed-batch cultures [J]. Biotechnol Lett, 2024, 46(5): 827-842. |
| [15] | 侯文义, 李相前. 酵母蛋白新食品原料研究进展及应用展望 [J]. 工业微生物, 2024, 54 (4): 51-56. |
| Hou WY, Li XQ. Research progress and application prospects of yeast protein as a novel food ingredient [J]. Industrial Microbiology, 2024, 54 (4): 51-56. | |
| [16] | Aggelopoulos T, Katsieris K, Bekatorou A, et al. Solid state fermentation of food waste mixtures for single cell protein, aroma volatiles and fat production [J]. Food Chem, 2014, 145: 710-716. |
| [17] | Duarte LC, Carvalheiro F, Lopes S, et al. Yeast biomass production in brewery’s spent grains hemicellulosic hydrolyzate [J]. Appl Biochem Biotechnol, 2008, 148(1/2/3): 119-129. |
| [18] | Yadav JSS, Bezawada J, Ajila CM, et al. Mixed culture of Kluyveromyces marxianus and Candida krusei for single-cell protein production and organic load removal from whey [J]. Bioresour Technol, 2014, 164: 119-127. |
| [19] | Yang R, Chen Z, Hu P, et al. Two-stage fermentation enhanced single-cell protein production by Yarrowia lipolytica from food waste [J]. Bioresour Technol, 2022, 361: 127677. |
| [20] | Liu BN, Song JZ, Li Y, et al. Towards industrially feasible treatment of potato starch processing waste by mixed cultures [J]. Appl Biochem Biotechnol, 2013, 171(4): 1001-1010. |
| [21] | Gao L, Meng J, Dai WL, et al. Deciphering cell wall sensors enabling the construction of robust P. pastoris for single-cell protein production [J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 178. |
| [22] | Johnson EA. Biotechnology of non-Saccharomyces yeasts—the ascomycetes [J]. Appl Microbiol Biotechnol, 2013, 97(2): 503-517. |
| [23] | 姜岷, 章文明, 马江锋,等. 一株利用甲醇制备高赖氨酸单细胞蛋白的多形汉逊酵母菌及其应用: CN105861343B [P]. 2019-05-07. |
| Jiang M, Zhang WM, Ma JF, et al. Application of Hansenula polymorpha yeast strain for preparing high lysine single-cell protein using methanol: CN105861343B [P]. 2019-05-07. | |
| [24] | Gorter de Vries AR, Pronk JT, Daran JG. Lager-brewing yeasts in the era of modern genetics [J]. FEMS Yeast Res, 2019, 19(7): foz063. |
| [25] | Stanley D, Fraser S, Chambers PJ, et al. Generation and characterisation of stable ethanol-tolerant mutants of Saccharomyces cerevisiae [J]. J Ind Microbiol Biotechnol, 2010, 37(2): 139-149. |
| [26] | Yu ZM, Zhao HF, Li HP, et al. Selection of Saccharomyces pastorianus variants with improved fermentation performance under very high gravity wort conditions [J]. Biotechnol Lett, 2012, 34(2): 365-370. |
| [27] | James TC, Usher J, Campbell S, et al. Lager yeasts possess dynamic genomes that undergo rearrangements and gene amplification in response to stress [J]. Curr Genet, 2008, 53(3): 139-152. |
| [28] | Shen N, Wang JJ, Liu CF, et al. Domesticating brewing yeast for decreasing acetaldehyde production and improving beer flavor stability [J]. Eur Food Res Technol, 2014, 238(3): 347-355. |
| [29] | Chen YF, Yang X, Zhang SJ, et al. Development of Saccharomyces cerevisiae producing higher levels of sulfur dioxide and glutathione to improve beer flavor stability [J]. Appl Biochem Biotechnol, 2012, 166(2): 402-413. |
| [30] | Liu XF, Wang ZY, Wang JJ, et al. Expression of GAI gene and disruption of PEP4 gene in an industrial brewer’s yeast strain [J]. Lett Appl Microbiol, 2009, 49(1): 117-123. |
| [31] | Brochado AR, Matos C, Møller BL, et al. Improved vanillin production in baker’s yeast through in silico design [J]. Microb Cell Fact, 2010, 9: 84. |
| [32] | Lee DN, Lloyd NDR, Pretorius IS, et al. Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion [J]. Microb Cell Fact, 2016, 15: 49. |
| [33] | Xue JK, Zhou JW, Li JH, et al. Systematic engineering of Saccharomyces cerevisiae for efficient synthesis of hemoglobins and myoglobins [J]. Bioresour Technol, 2023, 370: 128556. |
| [34] | Blieck L, Toye G, Dumortier F, et al. Isolation and characterization of brewer’s yeast variants with improved fermentation performance under high-gravity conditions [J]. Appl Environ Microbiol, 2007, 73(3): 815-824. |
| [35] | Huuskonen A, Markkula T, Vidgren V, et al. Selection from industrial lager yeast strains of variants with improved fermentation performance in very-high-gravity worts [J]. Appl Environ Microbiol, 2010, 76(5): 1563-1573. |
| [36] | Hansen J, Kielland-Brandt MC. Inactivation of MET10 in brewer’s yeast specifically increases SO2 formation during beer production [J]. Nat Biotechnol, 1996, 14(11): 1587-1591. |
| [37] | Yoshida S, Imoto J, Minato T, et al. Development of bottom-fermenting Saccharomyces strains that produce high SO2 levels, using integrated metabolome and transcriptome analysis [J]. Appl Environ Microbiol, 2008, 74(9): 2787-2796. |
| [38] | Vidgren V, Huuskonen A, Virtanen H, et al. Improved fermentation performance of a lager yeast after repair of its AGT1 maltose and maltotriose transporter genes [J]. Appl Environ Microbiol, 2009, 75(8): 2333-2345. |
| [39] | Voordeckers K, Kominek J, Das A, et al. Adaptation to high ethanol reveals complex evolutionary pathways [J]. PLoS Genet, 2015, 11(11): e1005635. |
| [40] | Govender P, Domingo JL, Bester MC, et al. Controlled expression of the dominant flocculation genes FLO1, FLO5 and FLO11 in Saccharomyces cerevisiae [J]. Appl Environ Microbiol, 2008, 74(19): 6041-6052. |
| [41] | Hansen EH, Møller BL, Kock GR, et al. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae) [J]. Appl Environ Microbiol, 2009, 75(9): 2765-2774. |
| [42] | Beekwilder J, van der Meer IM, Sibbesen O, et al. Microbial production of natural raspberry ketone [J]. Biotechnol J, 2007, 2(10): 1270-1279. |
| [43] | Yu F, Zhao XR, Zhou JW, et al. Biosynthesis of high-active hemoproteins by the efficient heme-supply Pichia pastoris chassis [J]. Adv Sci, 2023, 10(30): e2302826. |
| [44] | Shao YR, Xue CL, Liu WQ, et al. High-level secretory production of leghemoglobin in Pichia pastoris through enhanced globin expression and heme biosynthesis [J]. Bioresour Technol, 2022, 363: 127884. |
| [45] | Flight MH, Tait J, Chronopoulos T, et al. Analysing responsible innovation along a value chain-a single-cell protein case study [J]. Eng Biol, 2024, 8(1): 16-29. |
| [46] | Hadi J, Brightwell G. Safety of alternative proteins: technological, environmental and regulatory aspects of cultured meat, plant-based meat, insect protein and single-cell protein [J]. Foods, 2021, 10(6): 1226. |
| [47] | 王宇灵, 覃瑞, 刘虹, 等. 单细胞蛋白应用于食品工业的现状和展望 [J]. 中国食物与营养, 2019, 25 (10): 29-32. |
| Wang YL, Qin R, Liu H, et al. Current status and prospects of single cell protein application in food industry [J]. Food and Nutrition in China, 2019, 25 (10): 29-32. | |
| [48] | Anupama, Ravindra P. Value-added food: single cell protein [J]. Biotechnol Adv, 2000, 18(6): 459-479. |
| [49] | Onyeaka H, Anumudu CK, Okpe C, et al. Single cell protein for foods and feeds: a review of trends [J]. Open Microbiol J, 2022, 16: e187428582206160. |
| [50] | Weber S, Grande PM, Blank LM, et al. Insights into cell wall disintegration of Chlorella vulgaris [J]. PLoS One, 2022, 17(1): e0262500. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
